Transcriptional regulation of metabolic flux: a C. elegans perspective
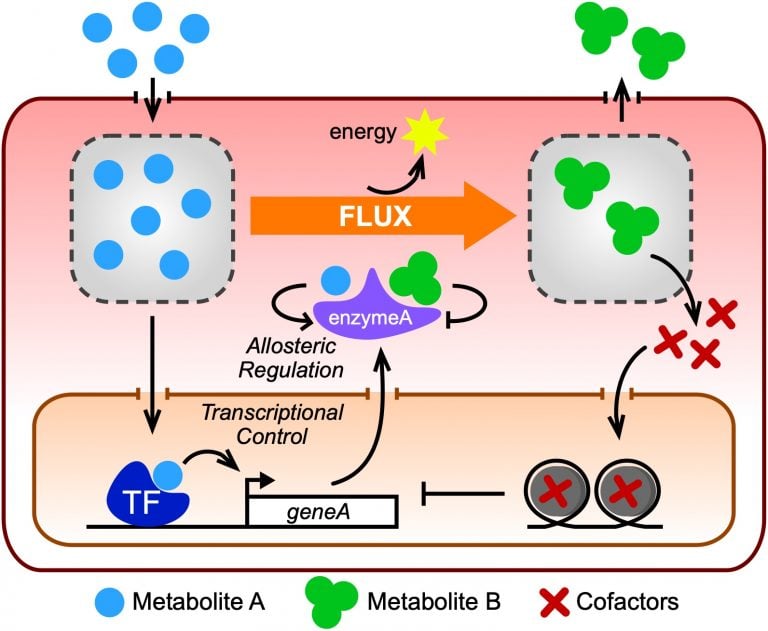 Metabolic reactions form the basis of life to generate biomass, produce energy and eliminate waste. Together, metabolic reactions function in the context of a highly interconnected metabolic network. Flux through this network needs to be adaptable depending on nutrient availability or metabolite buildup. Metabolic flux can be changed by different regulatory mechanisms, including the allosteric regulation of metabolic enzyme activity by small molecules, and by changes in metabolic gene expression. For the transcriptional control of metabolic genes to happen accurately, the metabolic status of the system needs to be relayed to the regulators of gene expression, such as transcription factors (TFs). While allosteric regulation of metabolic enzymes can only be studied for individual proteins, the transcriptional control of metabolism can increasingly be examined at a systems, or network level. Here, we discuss recent studies regarding the interplay between metabolic and gene regulatory networks using the nematode Caenorhabditis elegans. We discuss why C. elegans provides an elegant model system for studying the transcriptional regulation of metabolism and describe recent insights into its gene regulatory and metabolic networks. We then describe a recently discovered mechanism by which the buildup of a cellular metabolite can transcriptionally rewire metabolism. Finally, we discuss the future challenge of integrating large transcriptomic, proteomic and metabolomic datasets to fully understand the transcriptional regulation of metabolic flux.
Metabolic reactions form the basis of life to generate biomass, produce energy and eliminate waste. Together, metabolic reactions function in the context of a highly interconnected metabolic network. Flux through this network needs to be adaptable depending on nutrient availability or metabolite buildup. Metabolic flux can be changed by different regulatory mechanisms, including the allosteric regulation of metabolic enzyme activity by small molecules, and by changes in metabolic gene expression. For the transcriptional control of metabolic genes to happen accurately, the metabolic status of the system needs to be relayed to the regulators of gene expression, such as transcription factors (TFs). While allosteric regulation of metabolic enzymes can only be studied for individual proteins, the transcriptional control of metabolism can increasingly be examined at a systems, or network level. Here, we discuss recent studies regarding the interplay between metabolic and gene regulatory networks using the nematode Caenorhabditis elegans. We discuss why C. elegans provides an elegant model system for studying the transcriptional regulation of metabolism and describe recent insights into its gene regulatory and metabolic networks. We then describe a recently discovered mechanism by which the buildup of a cellular metabolite can transcriptionally rewire metabolism. Finally, we discuss the future challenge of integrating large transcriptomic, proteomic and metabolomic datasets to fully understand the transcriptional regulation of metabolic flux.
Giese GE, Nanda S, Holdorf AD, Walhout AJM. (2019) Transcriptional regulation of metabolic flux: a C. elegans perspective. Curr Opin Syst Biol 15, 12-18