Molecular Targeting of Transcriptional Factors by the Nedd4 Family of E3 Ubiquitin Ligase
Protein degradation plays a critical role in cell signaling. The stability of many regulatory proteins including transcriptional factors is under tight control for proper signal transduction. Selective degradation of proteins is carried out by the ubiquitin-proteasome pathway. Specificity of this pathway is largely determined by E3 ubiquitin ligases. There are two classes of E3 ligase: HECT and RING. While the RING E3 ligases transfer ubiquitin from E2 directly to protein substrates, the HECT E3 ligases form a thiol ester bond with ubiquitin before transferring ubiquitin to protein substrates. We study the Nedd4 family of HECT E3 ligase, which has nine members in mammals. They share similar structure features: N-terminal C2 domain, C-terminal catalytic HECT domain and 2-4 WW domains in the middle to mediate protein-protein interaction with substrates. WW domain is known to interact with PPXY motif, yet how such interaction determines substrate specificity is not clear. PPXY motif is found in many transcription factors, including those involved in TGF-β signaling, Wnt signaling, and polycomb complex. Using an unbiased genetic screen, we are interested in identifying E3 ligases that are responsible for the ubiquitination of these important transcriptional factors, and the specificity of substrate recognition by different members of the Nedd4 family of E3 ligase. Furthermore, the consequence of dysregulated ubiquitination of these transcriptional factors in diseases including cancer will be investigated.
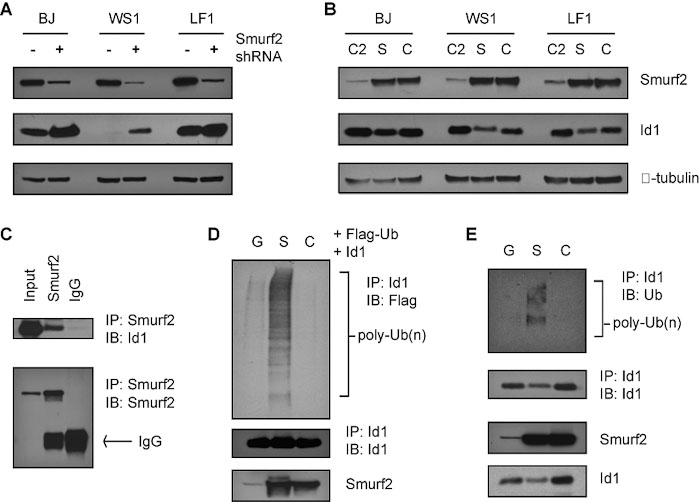
Smurf2 mediates ubiquitination of transcriptional regulator Id1. (A) Down-regulation of Smurf2 stabilizes Id1. (B) Ectopic expression of Smurf2 leads to decreased level of Id1. C2: Smurf2 C2 domain; S: Smurf2; C: ligase mutant C716A. (C) Smurf2 forms a complex with Id1. (D) Smurf2 induces ubiquitination of Id1. G: GFP; S: Smurf2; C: ligase mutant C716A. IP: immunoprecipitation. IB: immunoblot.
