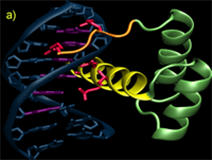
Homeodomains
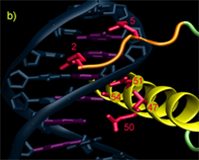
Homedomains are the second most common sequence-specific transcription factor in the genomes of higher eukaryotes. The simple DNA-binding architecture of the homeodomain, which consists of approximately 60 amino acids, facilitates the comparison of multiple factors with different specificity. The HD motif folds into a stable 3-helix bundle and recognizes 5 to 7 basepairs of DNA by positioning a single helix in the major groove and a flexible N-terminal arm over the minor groove (Figure A). Within this diverse family there is significant variation in the sequence composition of the homeodomain; for example the two major classes of homeodomains, typical and atypical, share on average only about 40% sequence identity and members from these two classes can recognize dramatically different DNA sequences. Nonetheless, the docking of HDs from these two classes with the DNA is nearly identical. This conserved binding geometry, most likely facilitated by common sets of contacts to the phosphodiester backbone, allows differences in amino acid sequence and DNA-binding specificity for various HDs to be interpreted within a common recognition framework. Most of the positions on this scaffold that are involved in sequence discrimination have been defined by structural, biochemical and mutagenic studies. Residues at positions 2, 3, 5, 6, 7 & 8 on the N-terminal arm and at positions 47, 50, 51, 54 & 55 on the recognition helix have been implicated as specificity determinants based on these studies (Figures B & C).
We have used the bacterial one-hybrid system to perform a comprehensive analysis of DNA-binding specificities for all 84 independent homeodomains in the genome of D. melanogaster in close collaboration with Mike Brodsky's laboratory. The homeodomain dataset represents the first complete description of DNA-binding specificities for any large family of TFs in any organism. In collaboration with Gary Stormo's laboratory, we have used this dataset to decipher new specificity determinants for this family of TFs that allows the semi-rational engineering of DNA-binding specficity. The homeodomain dataset has also allowed us to construct predictive models of DNA-bindign specificity for the homeodomain family, which has been elaborated into a web-based tool.
 |
 |
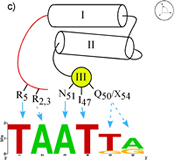 |
Legend for figures: DNA recognition by the homeodomain family. A) The interaction of Msx-1 (PDB ID: 1IG7) with its binding site is representative of homeodomain-DNA interactions. The homeodomain makes sequence-specific interactions with the DNA in both the major and minor groove. The N-terminal arm (orange) wraps around the 5' end of the DNA recognition sequence (magenta) while additional contacts are achieved in the major groove by the docking of a recognition helix (yellow) in the major groove nearly perpendicular to the DNA axis. Residues involved in base-specific interactions are shown (red). B) Close-up view of the DNA contacts observed in the Msx-1 structure, where residues at position 2 and 5 interact with bases in the minor groove and residues at positions 47, 50, 51 and 54 are positioned to make contacts in the major groove. C) Schematic overview of the key recognition contacts believed to be critical for DNA-sequence specificity in typical homeodomains. Positions on the N-terminal arm can influence the specificity at the first two positions of the binding site (shown as a sequence logo representation), where positions on the recognition helix affect specificity at the 3' end of the binding site.