Our research focus is on drug resistance.
x
We use multidisciplinary approaches, combining crystallography, enzymology, molecular dynamics and organic chemistry, to elucidate the molecular mechanisms of drug resistance.
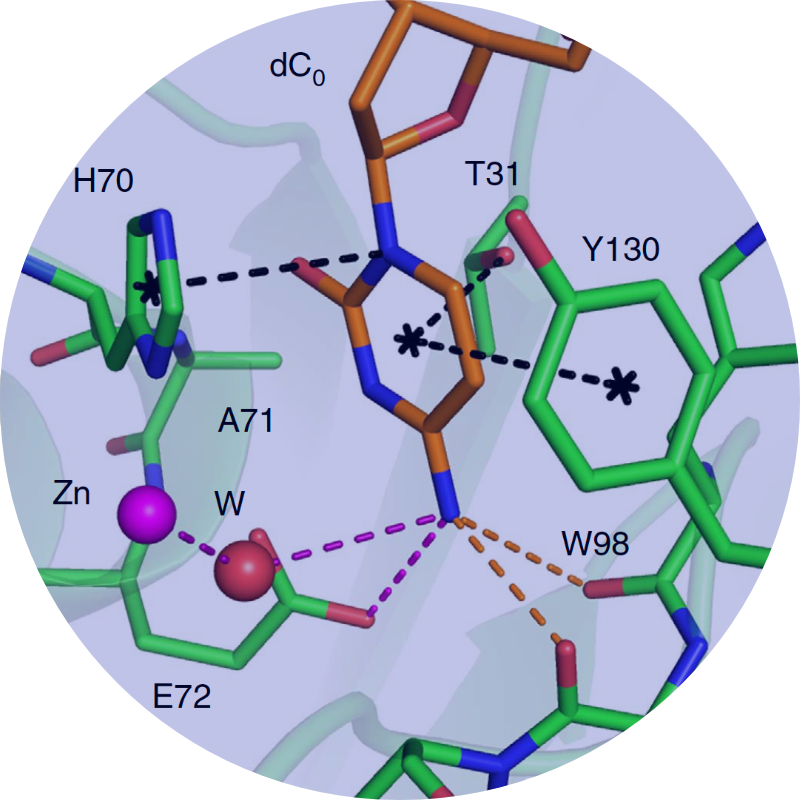
Resistance occurs when a heterogeneous populations of a drug target is challenged by the selective pressure of a drug. In cancer and viruses this heterogeneity is partially caused APOBEC3’s.
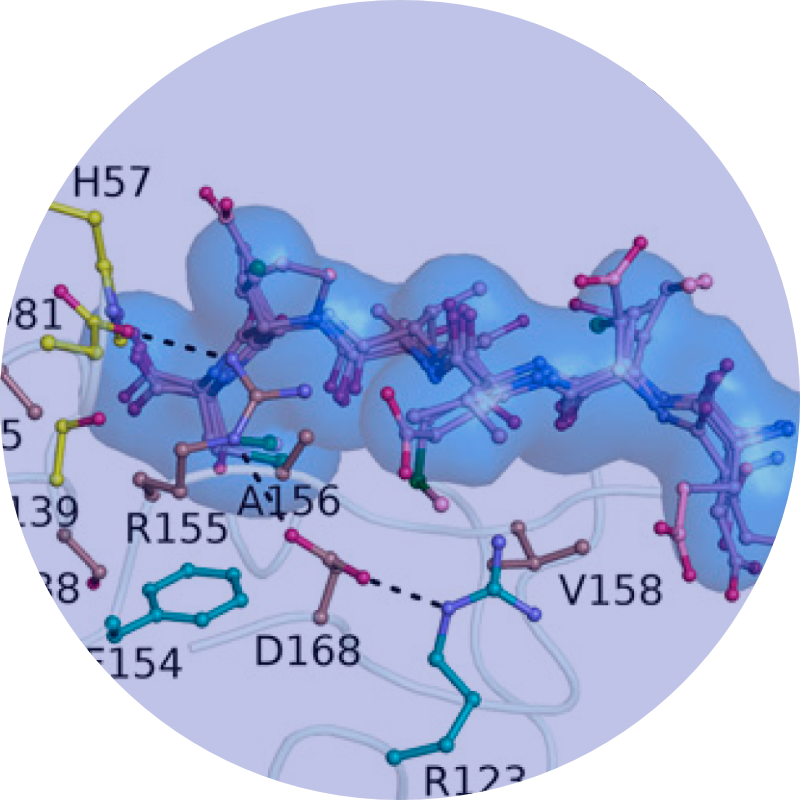
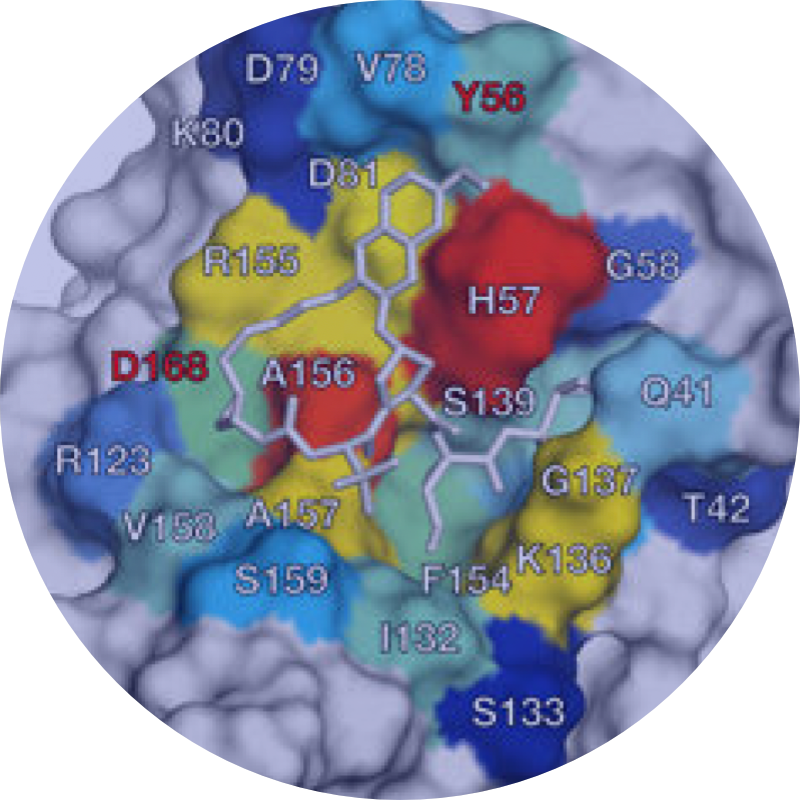
We discovered resistance mutations occur either where drugs physically contact regions of the drug target that are not essential for substrate recognition or alter the ensemble dynamics of the drug target favoring substrate.
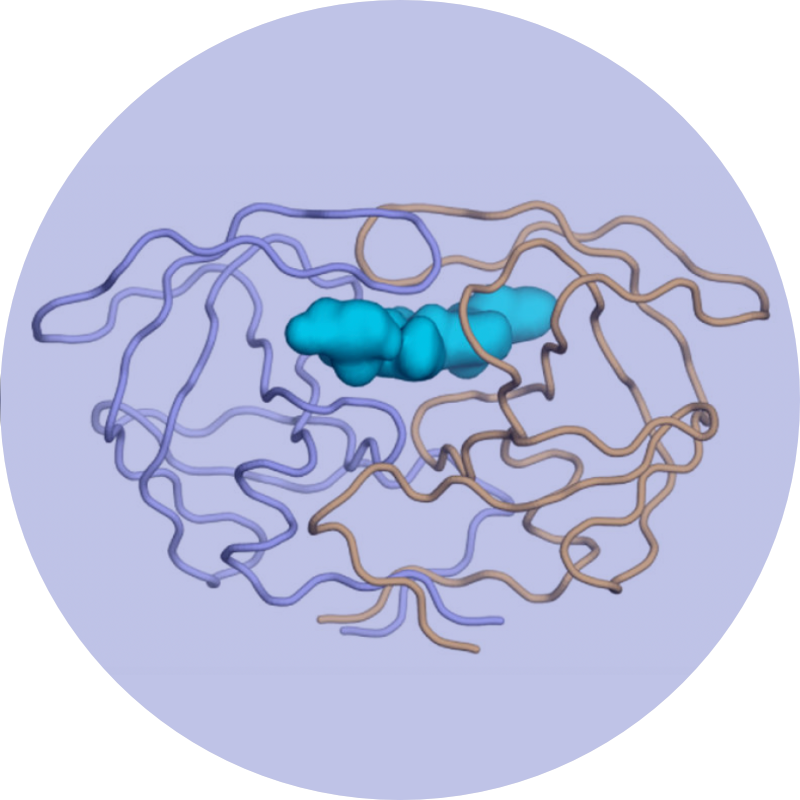
We leverage these insights into a new strategies in structure-based drug design to minimize the likelihood for resistance by designing inhibitors to stay within the substrate envelope.
This strategy not only describes most of the primary drug resistance for HIV, Hepatitis C viral protease inhibitors and influenza neuraminidase, but is generally applicable in the development of novel drugs that are less susceptible to resistance.
Research Areas
Our Approach
We take a problem-driven, multi-disciplinary and multi-technique approach to decipher the molecular underpinnings and mechanisms leading to drug resistance. We strive to apply a synergistic combination of various experimental and computational methods, and establish key collaborations with experts in other fields. We heavily rely on:
- Structural biology –having solved hundreds of crystal structures
- Parallel molecular dynamics simulations
- Structure based drug design
- Organic synthesis
- Enzyme inhibition and activity assays
- Deep sequencing data
Our unique integrative approach also aims to combine data from complementary techniques to effectively elucidate interdependency of molecular recognition.
Molecular illustrations on this webpage were generated by Leonora Martínez-Núñez, PhD.