High-throughput Phenotypic Profiling
|
Current deep sequencing technology allows the composition of complex libraries of >50,000 random transposon mutants to be quantitatively assessed. Using this approach, we are able to simultaneously determine the relative fitness of every viable loss of function mutant under specific selective conditions. We are using this strategy to comprehensively identify the bacterial genes required for adapting to different host microenvironments and to assign these genes to functional pathways. |
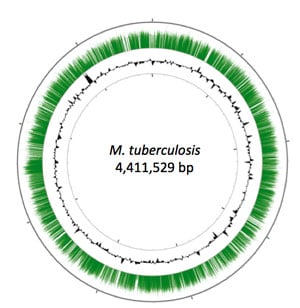 Depiction of the M. tuberculosis chromosome. Green bars represent the relative representation of mutants harboring transposon insertions at the indicated position. (See. Griffin et al. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathogens. 7(9): e1002251 (2011) |
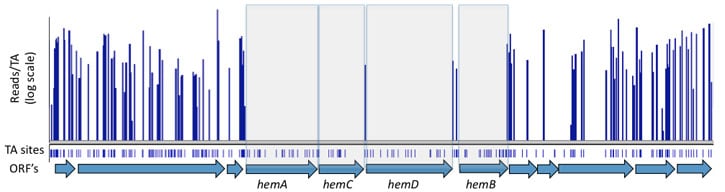
Gaps in transposon coverage identify essential chromosomal regions. Transposon insertions (blue bars) are not detected in heme biosynthetic genes that are essential for viability.
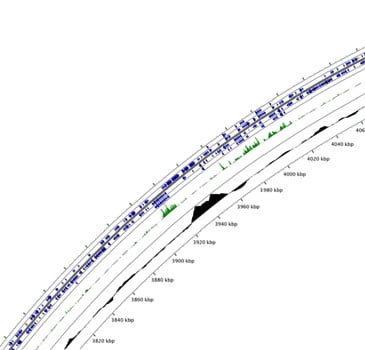
Comparitive analysis is used to identify conditionally essential genes. Genomic region encoding cholesterol catabolic genes is shown. Green bars indicate differential fitness of mutants in glycerol and cholesterol media.