Time-resolved crystallographic analysis of cooperative ligand binding
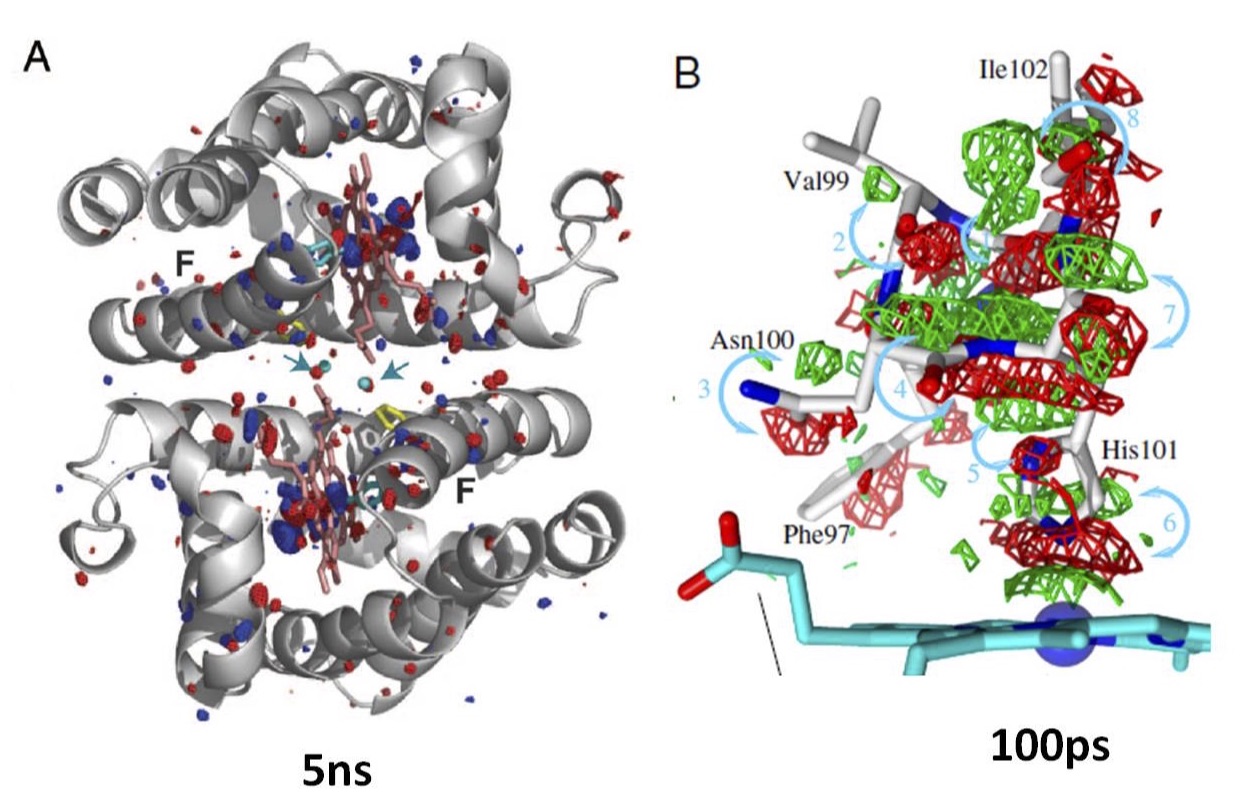
Invertebrate hemoglobins provide a number of useful model systems for exploring how protein subunits communicate. We have investigated systems ranging from the simplest possible allosteric systems to much more complex protein assemblages. As part of our efforts, we determined crystal structures of respiratory proteins with molecular masses of 3.6 x 106 Da comprising 180 different subunits. (Our crystal structure of Lumbricus erythrocruorin was highlighted as the March, 2013 molecule of the month at the protein data bank.) Our current efforts are focused on applying sub-ns time-resolved crystallographic analysis to a dimeric hemoglobin (Scapharca HbI) to dissect the time-dependent ligand-linked structural changes that underlie cooperative ligand binding. These experiments are being carried out at BioCARS (Advanced Photon Source) in collaboration with Dr. Vukica Srajer and Dr. Zhong Ren. They have already led to a new understanding of the role of water molecules in intersubunit communication as well as permitting the development of a novel dynamic model that incorporates a direct linkage between two active sites. In collaboration with Dr. Francesca Massi (BMP), we are combining the crystallographic results with NMR experiments that investigate the role of interface dynamics in cooperative behavior.