Paternal Effect Epigenetics
The past few decades have seen an important expansion of our understanding of inheritance, as a wide variety of epigenetically-inherited traits have been described. One implication of epigenetic inheritance systems is that they provide a potential mechanism by which parents could transfer information to their offspring about the environment they experienced, which, under certain environmental regimes, can in theory be adaptive. We have a longstanding interest in the question of whether environmental information can be passed on to offspring in mammals. We focus on paternal effects in mouse, as maternal environment can affect offspring both via epigenetic information in the oocyte, as well as direct impacts on gestating offspring (as in, eg, fetal alcohol syndrome).
Paternal effect paradigms
We have developed several paradigms in the lab in which male littermates are split to two conditions – Control and Low Protein diet, for example – then raised to sexual maturity and mated to Control females (Figure 1). Offspring phenotypes are characterized to determine whether the paternal environment has any impact on offspring physiology. We have a longstanding interest in paternal dietary effects – in our primary experimental paradigm, we find that male mice consuming a low protein diet (from weaning to sexual maturity) fathered offspring with decreased hepatic levels of cholesterol esters, and altered hepatic expression of lipid/cholesterol biosynthesis genes (Carone et al, Cell 2010). Similar results linking paternal diet to offspring metabolism have been reported by other groups including those of Morris, Patti, Bale, Lane, Chen, Ferguson-Smith and others, with many studies making the compelling case that a male’s diet, either during development or in adulthood, can affect a variety of phenotypes in his children.
More recently, in collaboration with Andrew Tapper’s laboratory, we have developed a paternal effect paradigm based on nicotine exposure. Here, we find that offspring of nicotine-exposed males exhibit enhanced resistance to toxic levels of both nicotine and cocaine (Vallaster et al, eLife 2017). We believe this paradigm is a major contribution to the field, as the use of a paternal perturbation with a defined ligand-receptor interaction enables us to use the tools of pharmacology to interrogate the specificity of the offspring response. Although the majority of our mechanistic studies (below) have been performed in the low protein system, we anticipate taking the same approaches in the nicotine system in the near future.
Paternal environmental effects on the sperm epigenome
It is a natural hypothesis that paternal environmental effects are transmitted via changes in one of the several sperm “epigenomes” (Figure 1).Study of the mechanisms underlying imprinting, PEV, epivariation in plants, and other epigenetic phenomena have uncovered three major classes of potential epigenetic information carrier – cytosine methylation, chromatin structure, and RNA. We have ongoing studies focused on all three of these epigenetic information carriers – see Carone et al Dev Cell 2014, Shea et al Dev Cell 2015, and Sharma et al Science 2016 for some of our early results on the sperm epigenome.
Post-testicular maturation shapes the RNA payload of sperm
During our investigations into dietary effects on the RNA payload of sperm, we discovered a surprising requirement for post-testicular sperm maturation in the biogenesis of small RNAs in sperm. After mature spermatozoa exit the testis, they spend ~10 days transiting a long convoluted tube known as the epididymis. We have found that large numbers of small RNAs – primarily 5’ ends of tRNAs known as tRNA fragments, or “tRFs”, but also other small RNAs such as microRNAs – are synthesized in the epididymal epithelium and trafficked to maturing sperm in small vesicles known as “epididymosomes.” (Figure 2) Evidence for this hypothesis includes: 1) high levels of tRFs (>80% of all small RNAs) in sperm isolated from cauda epididymis, despite negligible tRF levels (~2%) is any purified testicular sperm population, 2) high levels of tRNA cleavage in purified caput and cauda epididymis epithelium, 3) sperm from proximal and distal epididymis gain segment-specific small RNAs, as for example caput sperm have already gained high levels of tRF-Glu-CTC and tRF-Gly-GCC, while tRF-Val-CAC is restricted to cauda epididymis and cauda sperm, 4) epididymosomes carry small RNA populations that are strongly correlated with the RNA payload being gained by epididymal sperm, 5) epididymosomes can deliver small RNAs to immature sperm in vitro, and 6) Cre-Lox methods for expression of “tracer” RNAs in the caput epididymis definitively show that cauda sperm carry RNAs produced in the epididymis (unpublished data).
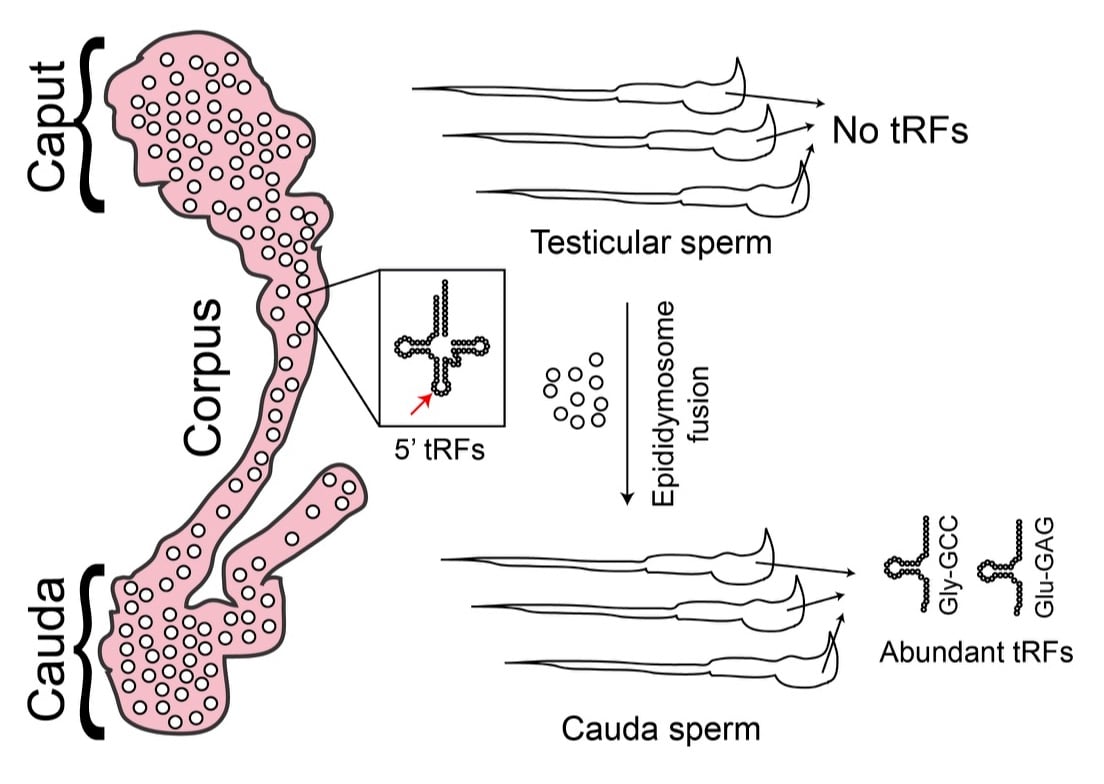
These data demonstrate a novel role for soma-germline communication in shaping the sperm RNA payload in mammals. Although the discovery of this paternal soma-germline communication pathway is in some ways quite surprising, it links mammalian spermatogenesis to many other gametogenesis systems in which somatic RNAs are communicated to germ cells via a variety of mechanisms. However, many features of this trafficking in mammalian sperm differ substantially from related pathways in other organisms, as for example small RNA shipment from soma to germline in ciliates and in Arabidopsis pollen occurs between nuclei sharing the same cytoplasm, whereas the trafficking step described here relies on vesicular transport. Our finding raises many questions about the molecular underpinnings of small RNA biogenesis, sorting, and trafficking throughout the epididymis.
Role of sperm-delivered small RNAs in preimplantation development
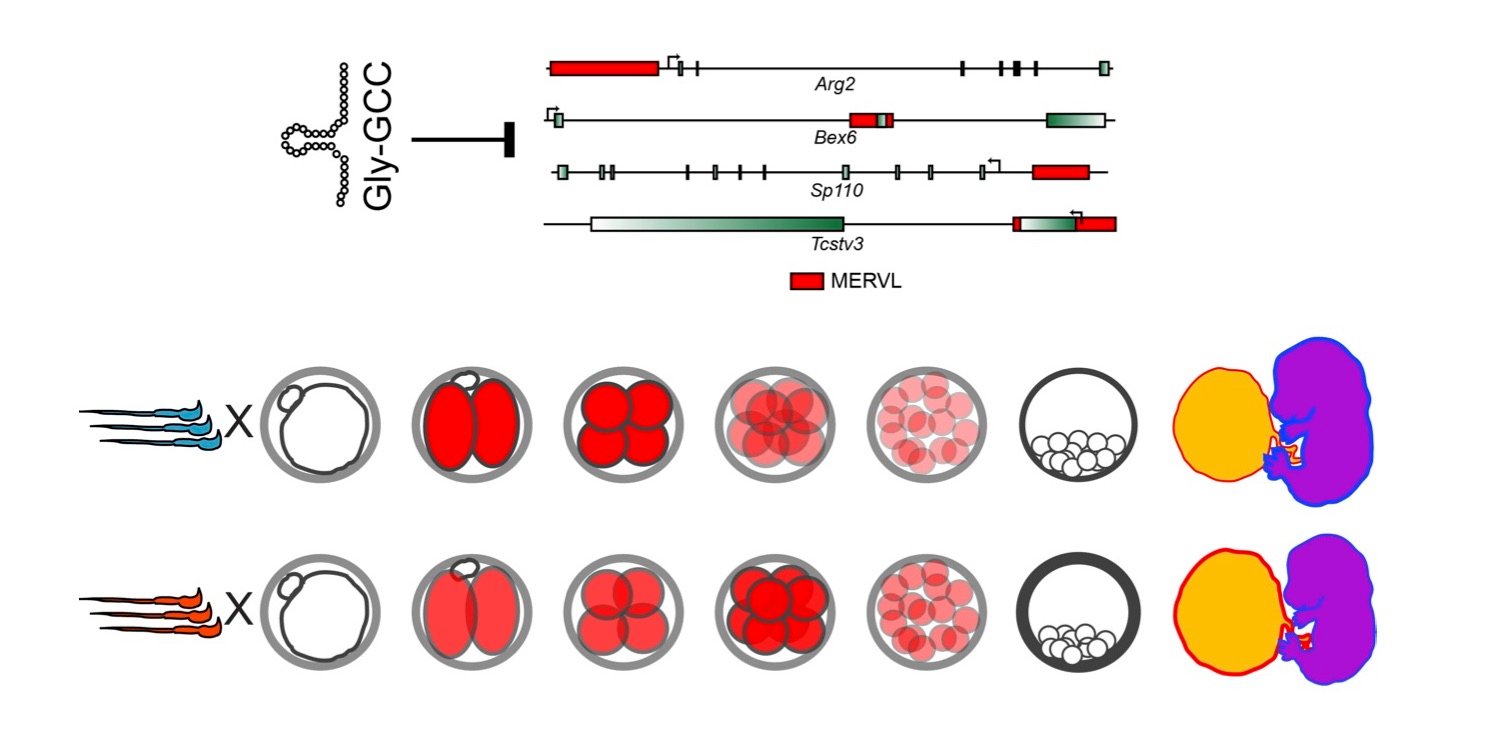
In our studies of paternal dietary effects, we found that levels of several tRNA fragments – most notably, fragments from several glycine isoacceptors – were increased in the sperm of Low Protein males. Detailed studies on tRF-Gly-GCC uncovered a surprising role for this small RNA in repression of genes associated with the long terminal repeats of the MERVL endogenous retroelement (Figure 3). This finding is particularly exciting given the deep ancestral relationship between tRNAs and LTR retroelements – tRNAs are almost universally used as the primers for reverse transcriptase in retroviruses and endogenous retroelements. However, surprisingly, our ongoing studies suggest that regulation of MERVL by tRF-Gly-GCC does not rely in any way on base pairing between the tRF and either gDNA or mRNA of the MERVL target genes. We are actively investigating the mechanistic basis for this regulation, and are interested in the regulatory roles of other tRFs.