Nuclear and Genome Organization in Development and Cancer
Normal somatic cells have highly compartmentalized nuclear structure |
|||
|
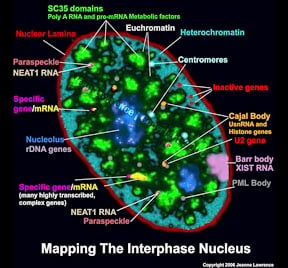
The functional complexity of the nucleus is reflected in its remarkable structural complexity. As seen in the image to the right, in addition to DNA, the somatic cell nucleus is comprised of a number of different domains and nuclear bodies, such as nucleoli and SC-35 speckles, which are involved in distinct aspects of gene expression. These non-membrane bound compartments form by liquid phase separation, allowing molecules within the relatively stable structures to remain dynamic. Many of these nuclear compartments contain unique noncoding RNAs and often are associated with specific gene loci |
|
||
Nuclear Speckles (SC-35 Domains) |
|||
|
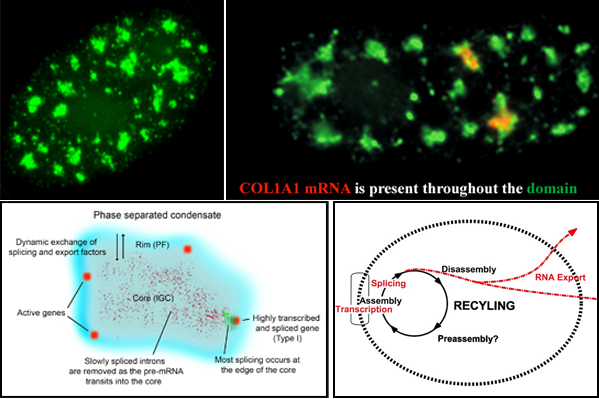
Nuclear speckles, also referred to as SC-35 domains, are nuclear structures that are enriched with factors involved in transcription, RNA processing and export, as well as ncRNAS such snRNAs and the lncRNA MALAT1 (also called NEAT2). Specific genes localize at the edge of speckles, and the concept that they are hubs for transcription, mRNA processing and export has been suggested by our lab since the mid-1990s. Now, genome wide studies have found that active genes throughout the genome associate with speckles. Speckles (top left image) are membraneless condensates that facilitate expression of associated genes through increased concentration and dynamic exchange of factors involved in mRNA metabolism. Top right image: Some speckles have Col1A1 RNA throutout them. Lower left model: Many active genes (red) position in the rim (blue region), which corresponds to perichromatin fibril (PF) ultrastructures, where transcription occurs, but are adjacent to the speckle core (corresponding to interchromatin granule cluster (IGC) ultrastructures), which contains transcription, splicing and export factors. All or almost all introns (green) are removed within the periphery at or near the gene, while some introns can be such as slow splicing introns of COL1A1 mRNA (punctate red signals) distribute throughout the core. Highly expressed, intron-rich genes such as Col1A1 always associate with speckles and their nascent mRNAs can be found within the speckle. We propose that these high demand genes nucleate formation of speckles with which other genes then associate to facilitate their own expression.Lower right: Model suggesting a functional rationale for coupling the completion of mRNA maturation and release for mRNA export with the recycling/preassembly of factors within speckles. Since pre-mRNA processing requires interaction of many different factors, their concentration in condensates would facilitate recycling and reuse for expression of adjacent genes. Selected Lawrence lab publications about SC-35 speckles: Molecular anatomy of a speckle. Anat Rec A Discov Mol Cell Evol Biol. 2006 Jul;288(7):664-75. A three-dimensional view of precursor messenger RNA metabolism within the mammalian nucleus. Science. 1993 Feb 26;259(5099):1330-5. |
|||
NEAT1 RNA is an architectural RNA that scaffolds a large nuclear structure |
|||
|
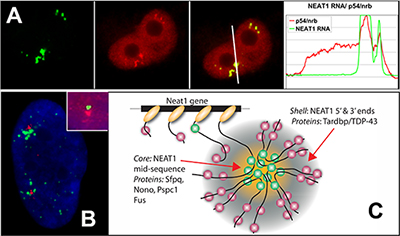
NEAT1 RNA is a noncoding RNA that strictly localizes in paraspeckles, ubiquitous nuclear structures (~10-30/nucleus) which abut the larger nuclear speckles, and provides the essential architectural scaffold for their formation. Paraspeckles can be found in all human primary and transformed cells and are enriched for over 40 different RNA binding proteins, such as PSP1. Paraspeckles are thought to sequester nuclear-retained RNAs and proteins involved in gene regulation or pri-miRNA processing.
References: An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009 Mar 27;33(6):717-26. |
|||
Pluripotent stem cells have a unique nuclear organization |
|||
|
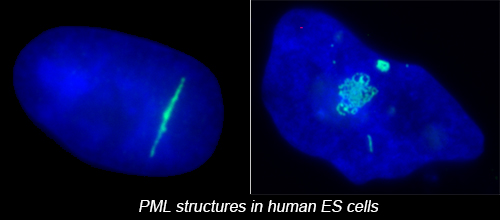
The complex nuclear structure of somatic cells is a key component of epigenomic regulation, yet little is known about nuclear structure and genomic organization of pluripotent cells (e.g. hESC). Here we surveyed several nuclear structures in pluripotent and transitioning hESC. We find hESC have unique nuclear organization and lack numerous defined structural compartments seen in somatic cells. They also exhibit dramatically different PML-defined structures, which in somatic cells are linked to gene regulation and cancer.
Reference: |
|||
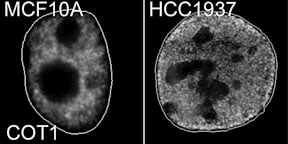
Cancer cells lose the peripheral heterochromatic compartment |
|||
Reference: |
|||
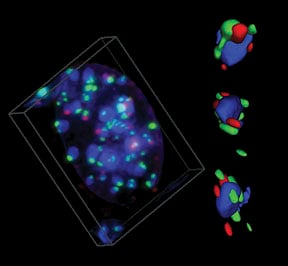
BRCA1 associates with centromeric satellite DNA during its replication |
|||
|
Proper replication of centromeric DNA is essential to ensure appropriate segregation of chromosomes at mitosis. The link between BRCA1 and centromeric replication could connect BRCA1 to both epigenetic and genetic instability in cancers. We suggest that heterochromatic instability is a common but largely unexplored mechanism, leading to widespread genomic misregulation and the evolution of some cancers.
Reference: |
|||