Genome editing tools
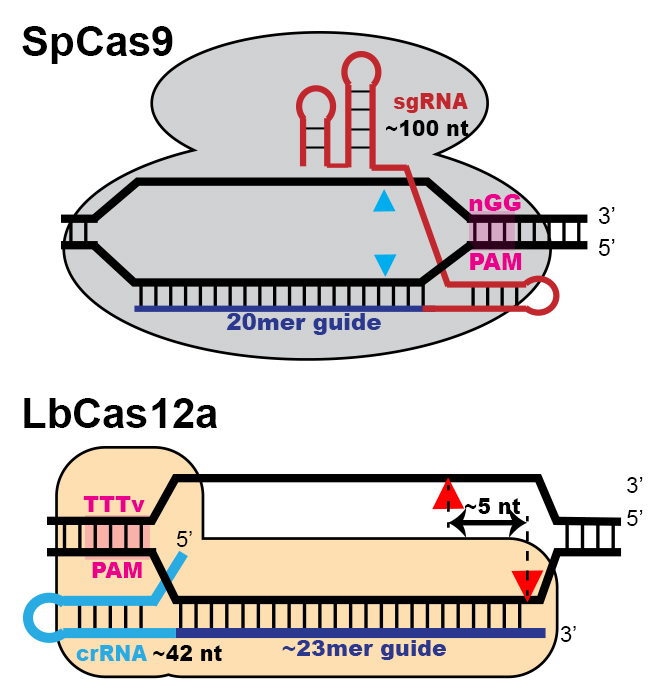 In 2008, together with the Wolfe Lab, we were among the first two groups to demonstrate the application of programmable nucleases for introducing targeted deletions into the zebrafish germline. Since that time, we have continued our collaborative efforts to further develop and optimize targeted strategies for manipulating the zebrafish genome. Our recent efforts have focused on Cas12a, an RNA-programmable nuclease similar to Cas9 (see Figure). However, unlike Cas9, the guide RNA for Cas12a is shorter and targets a distinct protospacer sequence (TTTV). Also, Cas12a leaves a single strand overhang following cleavage and the location of its PAM allows it to repeatedly cut imprecisely repaired edits. These distinctions suggest that Cas12a may yield improved homology-directed repair rates. Ongoing studies in the Lawson Lab are focused on optimizing Cas12a for this purpose.
In 2008, together with the Wolfe Lab, we were among the first two groups to demonstrate the application of programmable nucleases for introducing targeted deletions into the zebrafish germline. Since that time, we have continued our collaborative efforts to further develop and optimize targeted strategies for manipulating the zebrafish genome. Our recent efforts have focused on Cas12a, an RNA-programmable nuclease similar to Cas9 (see Figure). However, unlike Cas9, the guide RNA for Cas12a is shorter and targets a distinct protospacer sequence (TTTV). Also, Cas12a leaves a single strand overhang following cleavage and the location of its PAM allows it to repeatedly cut imprecisely repaired edits. These distinctions suggest that Cas12a may yield improved homology-directed repair rates. Ongoing studies in the Lawson Lab are focused on optimizing Cas12a for this purpose.
To see our most recent work on Cas12a, check out the paper below:
Liu, P., Luk, K., Shin, M., Idrizi, F., Kwok, S. Roscoe, B., Mintze, E., Suresh, S., Morrison, K., Frazao, J. B., Bolukbasi, M. F., Ponnienselvan, K., Luban, J., Zhu, L. J., Lawson, N. D., Wolfe, S. A. (2019) Enhanced Cas12a editing in mammalian cells and zebrafish. Nucleic Acids Research, on-line 3/20/2019.
If you want to try using Cas12a in zebrafish, check out our protocols page.