XIST RNA and X inactivation
XIST RNA coats the inactive X-chromosome and induces epigenetic silencing. |
|
| In order to dosage compensate X-linked genes between males and females, one of the two X-chromosomes in female cells is almost completely silenced. XIST RNA is a large non-coding RNA (~17kb mature transcript) that is transcribed exclusively from the inactive X-chromosome and “paints” the full length of the chromosome in cis (it never leaves the chromosome it is transcribed from and is not seen in the cytoplasm). XIST RNA then initiates a cascade of events that leads to the epigenetic silencing of one of the two X-chromosomes in most of the cells in female mammalian embryos. | |
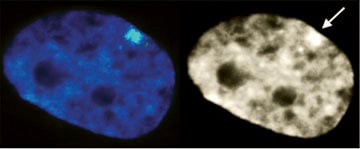 |
XIST RNA (green in left image) paints the inactive X-chromosome in interphase human female cells. |
|
|
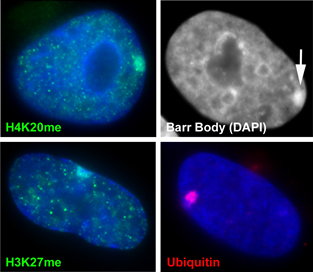
The inactive chromosome exhibits numerous epigenetic hallmarks. For example, methylation of histones H4K20 and H3K27, H2AK119 ubiquitination and condensation into a compact structure called the Barr body that stains densely with the DNA stain DAPI. |
Selected references:Hall LL, Lawrence JB. The cell biology of a novel chromosomal RNA: chromosome painting by XIST/Xist RNA initiates a remodeling cascade. Semin Cell Dev Biol. 2003 Dec;14(6):369-78. |
|
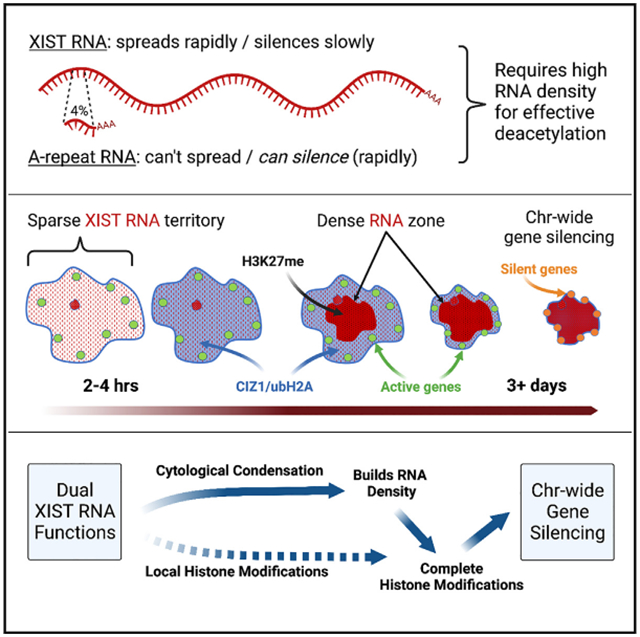
XIST lncRNA condenses cytoarchitecture before widespread gene silencing |
|
|
|
|
Reference: |
|
How the Barr Body is organized |
|
|
|
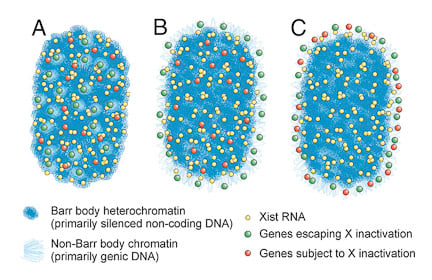
It was long believed that the heterochromatic Barr Body visible in the center of the inactive X territory was the manifestation of the silencing of the protein coding genes on the X-chromosome, and that silenced genes were located within this heterochromatic structure. However, it was unknown whether the genes that escaped inactivation were also within the Barr Body but resistant to its effects (model A), or were located outside the Barr Body (model B). We were surprised to find that all genes regardless of their activity were located outside the heterochromatic Barr Body! |
|
|
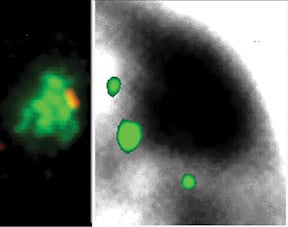
Genes are outside the Barr body.Left image: genes (red) are seen at the inner edge of the XIST RNA territory (green). Right image: A pool of 3 X-linked genes (small green dots) are located outside the Barr body (black). |
|
|
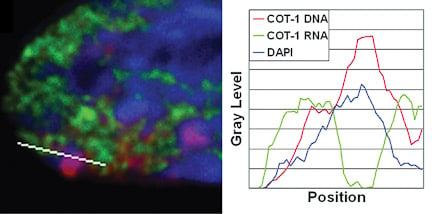
Repeat DNA is enriched in the Barr Body and is silencedIf genes are not within the Barr Body, then this then begs the question what is inside. By using the Cot-1 DNA fraction of the genome (all the highly repetitive sequences) as a probe for RNA/DNA FISH, we found that the Barr Body was made up of the repeat fraction of the chromosome, and that this DNA was transcriptionally silent. However, Cot-1 RNA was found transcribed throughout the rest of the euchromatic regions on the nucleus, and across the active X-chromosome territory. |
Select references: |
|
How XIST binds to chromatin |
|
|
Our previous studies have shown that the XIST RNA does not bind exclusively via DNA/RNA binding. If DNA is removed from a nucleus in situ (by DNAse), XIST RNA remains tightly bound to the remaining protein component of the cell. |
|
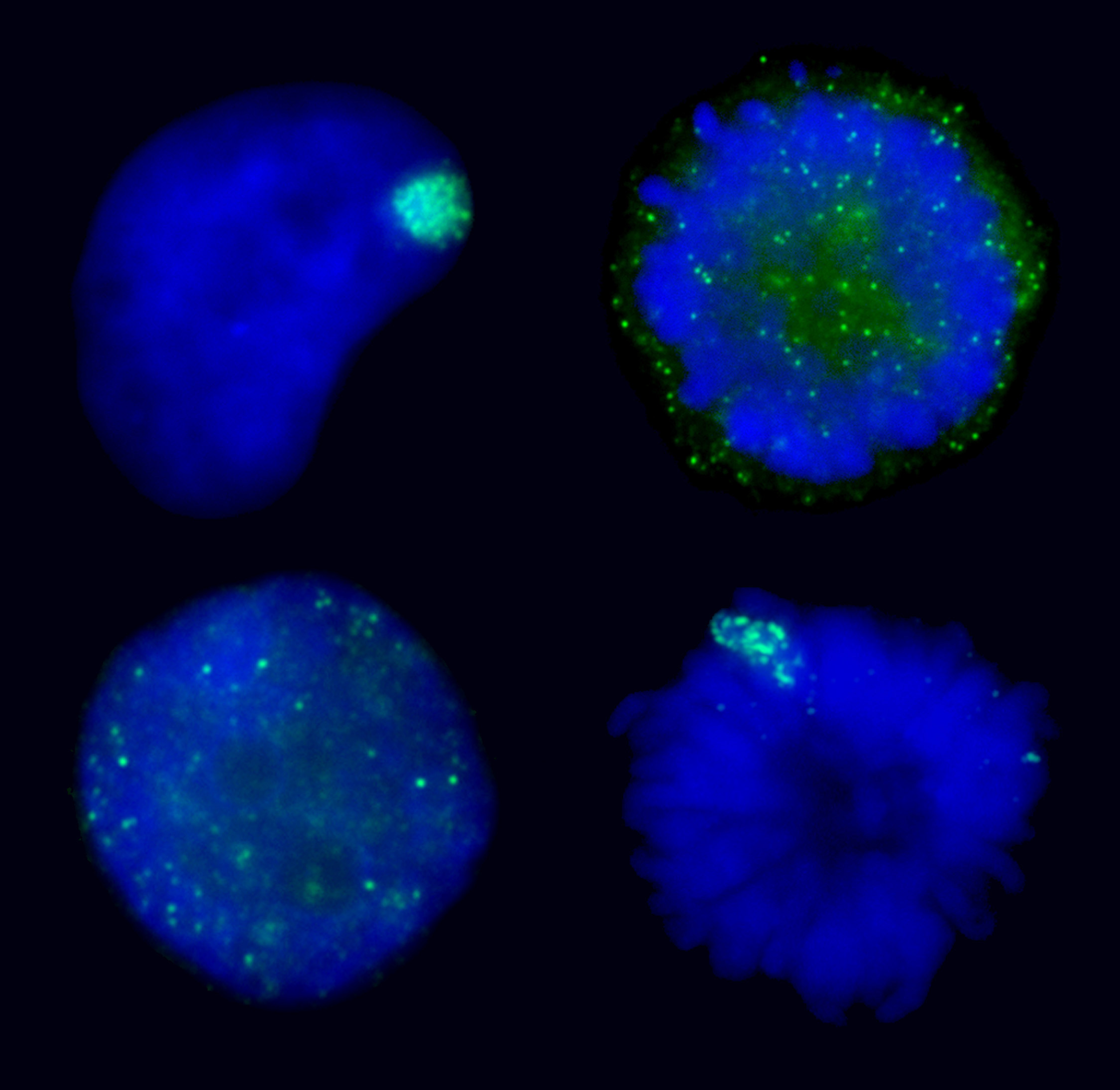
AURKB mediated chromatin modifications regulate XIST RNA binding.XIST RNA naturally falls off the inactive chromosome during mitosis, suggesting mitotic protein modifications may play a role. Here we find that manipulating the activity of the mitotic kinase Aurora B (AURKB) in interphase or mitosis can affect the binding of XIST to the chromosome. This suggests that XIST RNA binding to the chromosome is affected by phosphorylation of an AURKB substrate. The ability to manipulate XIST RNA behavior in vivo provides a strategy to determine which chromosome/ chromatin associated proteins most closely mirror the binding pattern of XIST RNA. |
Manipulating XIST RNA’s interaction with the chromosome: Top images show XIST RNA localization in normal interphase and its natural release at mitosis. Bottom two images show induced interphase release or induced metaphase retention of XIST RNA. |
XIST binding to chromatin is complex.
|
|
|
|
|
Select references: |
|

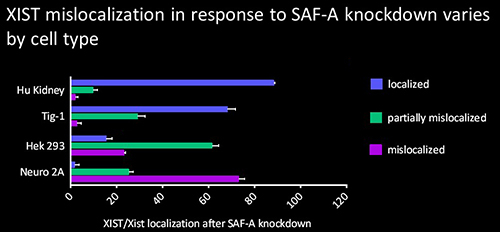 The protein SAF-A has been suggested to connect XIST to DNA through its RNA- and DNA-binding domains. We tested the role of SAF-A in localizing human XIST RNA in different cell types and found that it was generally not essential to maintain XIST RNA localization in normal cells such as Tig-1 fibroblasts and kidney cells whereas loss of SAF-A resulted in more mislocalization in transformed cells (HEK293, Neuro2A).
The protein SAF-A has been suggested to connect XIST to DNA through its RNA- and DNA-binding domains. We tested the role of SAF-A in localizing human XIST RNA in different cell types and found that it was generally not essential to maintain XIST RNA localization in normal cells such as Tig-1 fibroblasts and kidney cells whereas loss of SAF-A resulted in more mislocalization in transformed cells (HEK293, Neuro2A).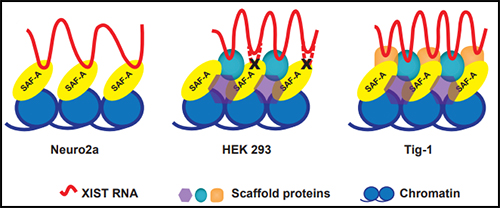 Our results suggest that SAF-A is one of multiple redundant anchors in primary somatic cells and that other anchors are often lost or compromised in many transformed cells. We also found that human SAF-A’s putative RNA-binding (RGG) domain was not required to maintain localized XIST RNA, further suggesting that XIST RNA is not simply tethered to chromatin by a single-molecule SAF-A “bridge.”
Our results suggest that SAF-A is one of multiple redundant anchors in primary somatic cells and that other anchors are often lost or compromised in many transformed cells. We also found that human SAF-A’s putative RNA-binding (RGG) domain was not required to maintain localized XIST RNA, further suggesting that XIST RNA is not simply tethered to chromatin by a single-molecule SAF-A “bridge.”