We strive to characterize molecular interactions within and between proteins, including their substrates, antibodies, and small molecule compounds using structural and computational biology. We leverage our thorough understanding of these molecular interactions to develop clinically relevant solutions, including designing and optimizing inhibitors.

Beyond our passion for fundamental molecular and structural biology, we are motivated by our goal of contributing to the development of new treatment options for various human diseases.
x
x

We synergistically utilize experimental and computational techniques to gain fundamental insights into molecular recognition events involving protein-protein and protein-ligand interactions. Much of our work focuses on developing design strategies for drug candidates against resistant viruses, but we also apply this framework to other diseases such as cancer and Type I Diabetes.
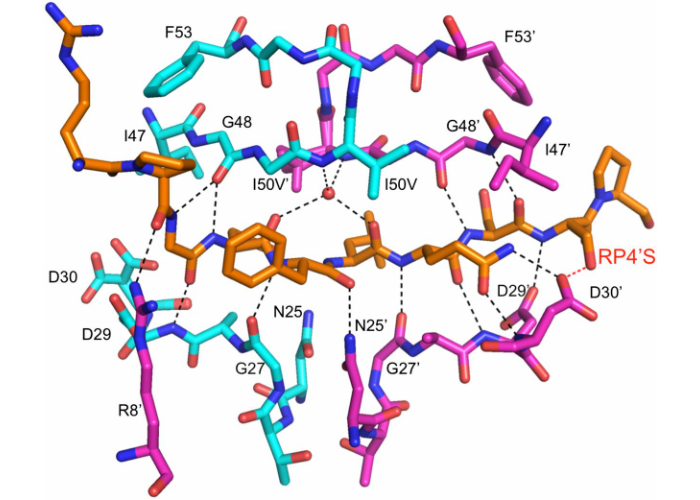 Structural basis and distal effects of Gag substrate coevolution in drug resistance to HIV-1 protease
Structural basis and distal effects of Gag substrate coevolution in drug resistance to HIV-1 protease
Ozen, A., Lin, K.H., Kurt Yilmaz, N*#., Schiffer, C.A.
*joint first author #corresponding author
Proc Natl Acad Sci U S A. 2014 Nov 11;111(45):15993-8. doi: 10.1073/pnas.1414063111. Epub 2014 Oct 29. PMID: 25355911; PMCID: PMC4234576.
While evolving high levels of resistance to protease inhibitors, HIV-1 accumulates mutations not only in the protease but also its substrates. In this work we revealed the molecular-level interplay between these two types of resistance mutations, and how co-evolution of the substrate restores the structure and dynamics of the complex to allow evolution of resistant viral variants.
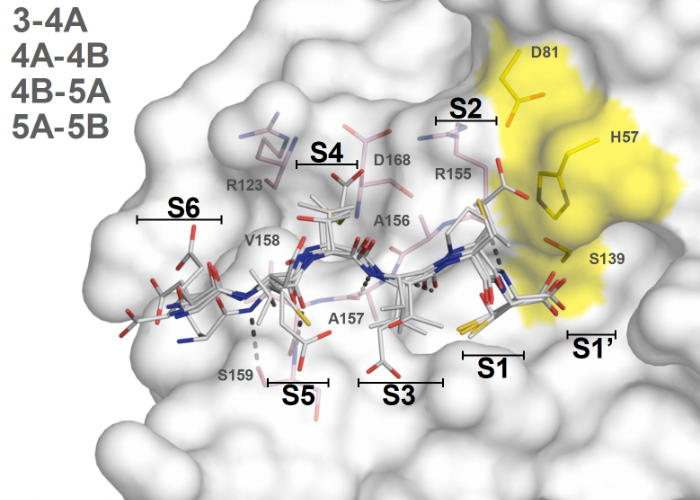 Avoiding drug resistance by substrate envelope guided design: toward potent and robust HCV NS3/4A protease inhibitors
Avoiding drug resistance by substrate envelope guided design: toward potent and robust HCV NS3/4A protease inhibitors
Matthew, A.N., Zephyr, J., Rao, D.N., Henes, M., Kamran, W., Kosovrasti, K., Hedger, A.K., Lockbaum, G.J., Ali, A., Kurt Yilmaz, N.*, Shiffer, C.A.*
*joint corresponding author
mBio. 2020 Mar 31;11(2):e00172-20. doi: 10.1128/mBio.00172-20. PMID: 32234812; PMCID: PMC7157764.
Leveraging our knowledge of the intermolecular interaction between HCV NS3/4A protease and its natural substrates, we designed improved inhibitors that display better potency against clinically relevant resistant variants of the virus. These lessons are transferable to the design of potent and robust inhibitors against other drug targets.
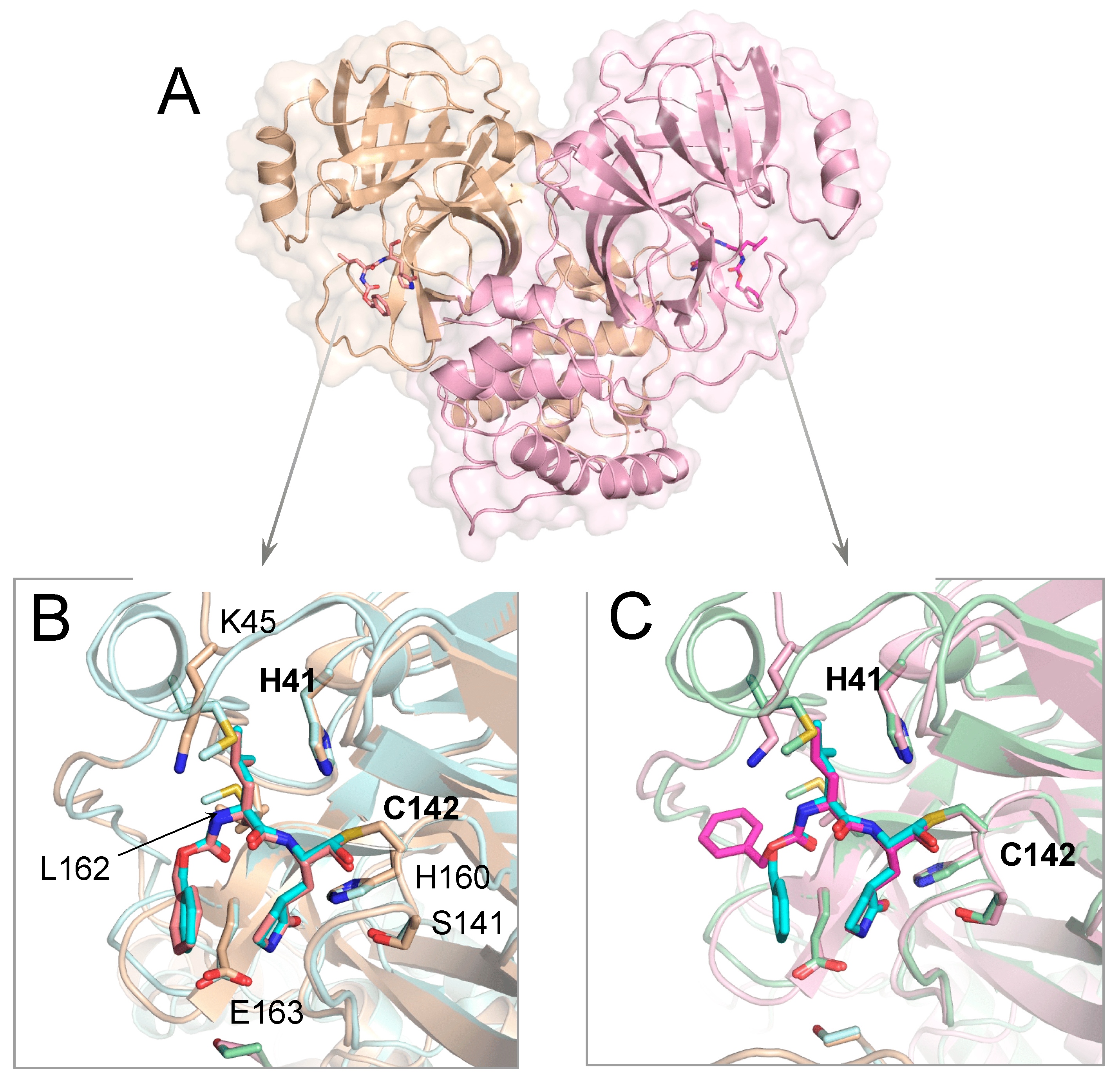 Crystal Structures of Inhibitor-Bound Main Protease for Delta-and Gamma Coronaviruses
Crystal Structures of Inhibitor-Bound Main Protease for Delta-and Gamma Coronaviruses
Zvornicanin SN, Shaqra AM, Huang QJ, Ornelas E, Moghe M, Knapp M, Moquin S, Dovala D, Schiffer CA, Kurt Yilmaz N
Viruses. 2023 Mar 18;15(3):781. doi: 10.3390/v15030781. PMID: 36992489; PMCID: PMC10059799.