Structural and Mechanistic Studies of Alternative Clamp Loaders
Eukaryotic organisms have an array of alternative clamp loaders that are used in pathways distinct from DNA replication, such as DNA damage and checkpoint control, sister chromatid cohesion, and recombination (see Kelch Biopolymers 2016). These clamp loaders have been suggested to be important targets for novel cancer therapeutics. Despite differing from the replicative clamp loader by just one subunit in the pentameric assembly, the alternative clamp loaders exhibit significant functional and mechanistic differences. For example the Rad24 clamp loader binds a different sliding clamp, the ’9-1-1 complex’ (Majka & Burgers, PNAS 2003), and functions at the 5’ DNA junction, the opposite polarity to that of the replicative clamp loader (Ellison & Stillman, PLOS Bio 2003).
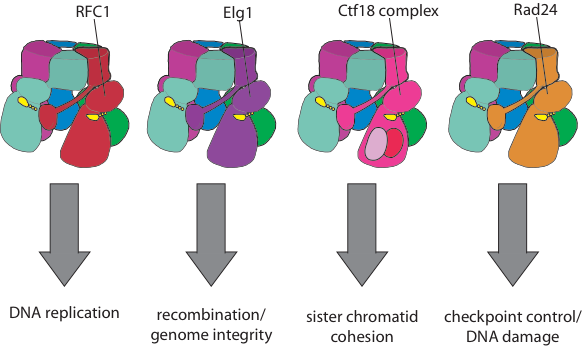
Our goal is to determine the structural and biochemical basis for the mechanistic and functional differences of the alternative clamp loaders from their replicative clamp loader cousins. We will also understand how alternative clamp loaders are regulated and integrated into the pathways of recombination, sister chromatid cohesion and DNA repair. These studies will reveal how AAA+ machines can be regulated, as well as identifying the mechanistic underpinnings of these key components of these important cellular pathways.

