Transcriptional Regulation of Signaling
Understanding how genes are regulated in various cell types and contexts is a major challenge in biology. With the advances in high-throughput assays it is now evident that genes exist in vast and complex regulatory landscapes of promoters and enhancers. In general promoters are thought to be acting as ON/OFF switches in cell type invariant manner, whereas enhancers in general, provide fine tuning and cell-type specificity. Genomewide maps of chromatin modifications, chromatin accessibility have revealed thousands of putative distal regulatory elements which contain DNA binding motifs. However, how the binding of a transcription factor to a promoter or enhancer affects target gene expression is still unclear. A second challenge in the study of regulatory elements is the sheer complexity of the regulatory architecture. Specifically, it has become clear that enhancer/gene interactions are complex, involving up to tens of different enhancers per gene. In addition, it has been shown that enhancer elements are likely to interact with more than one target gene.
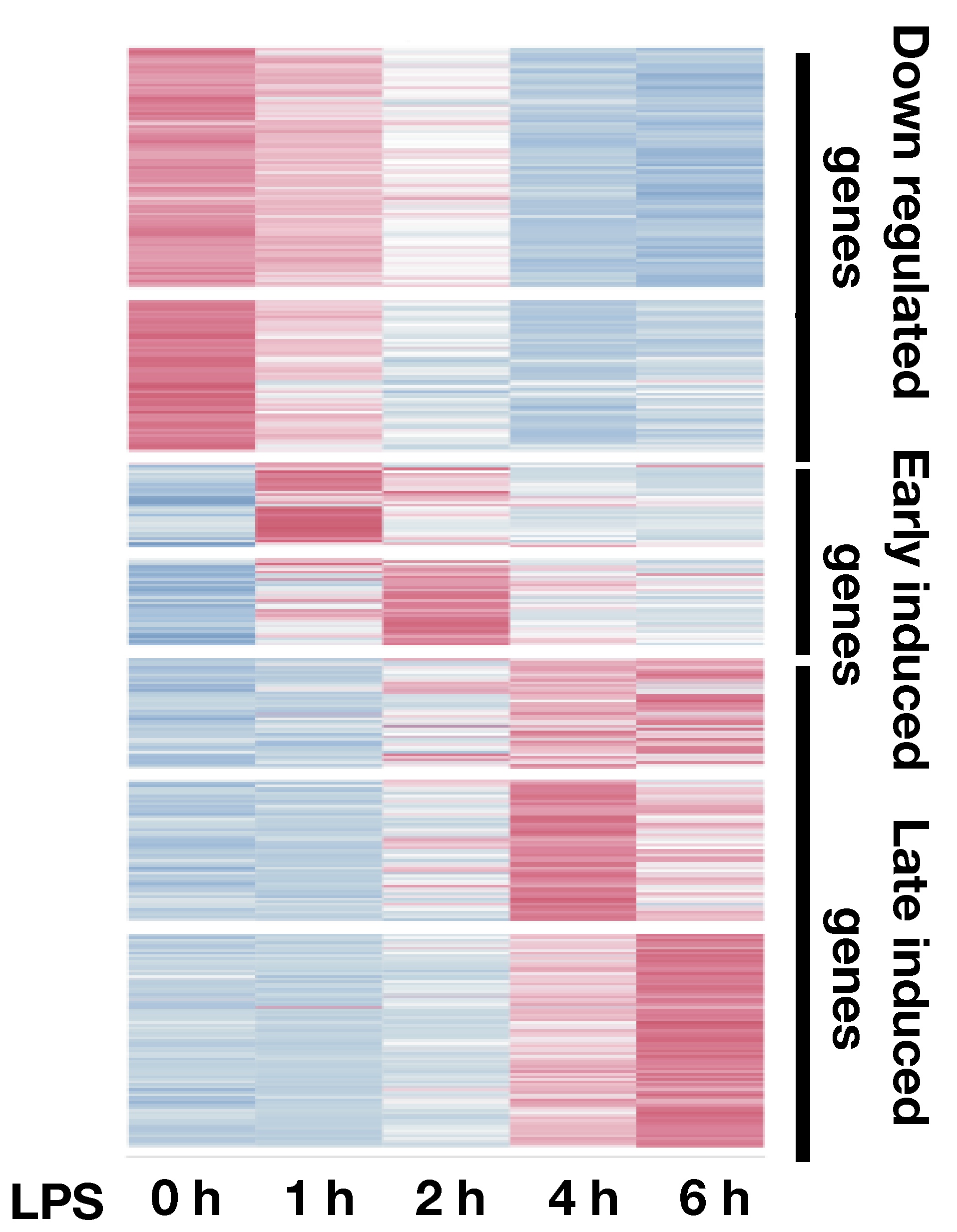
To understand the regulatory grammar of gene regulations, we use innate immune cells stimulated with TLR4 ligand as a model. This system has enabled us to map the steady state and context dependent cis-regulatory elements. Our integrated analysis of temporal datasets of transcription binding and gene expression showed that binding of different factors is responsible for subtle expression patterns that control specific pathways. These pathways have very distinct forms of regulation: a minority of pathways are regulated by few transcription factors (e.g. Stat1 and Stat2) whose targets are very responsive to knock down of these factors and have very conserved binding sites. In contrast, most pathways are controlled by a larger set of redundant transcription factors, whose binding has an additive effect and expression where the number rather than the type of factors bound gives rise to different expression levels. We also have shown that regulatory complexity, defined simply as the number of regulatory elements associated with a gene, is associated with important gene characteristics: low expression variance across evolution, activation of key genes that regulate cell fate decisions and rapid and high activation in signal transduction pathways. Our analysis also revealed that cis-regulatory elements that show temporal kinetics tend to associate with genes having similar transcriptional kinetics.
Our lab seeks to understand how different cell types use different combinations of regulatory elements to control cell type specific temporal expression kinetics. To crack the regulatory code of mammalian genes, we are integrating molecular evolution, population genetics, DNA conformation and high throughput functional genomics data.