Evolution of Cis Regulatory Elements
“The Human Genome Project (HGP) was the international, collaborative research program whose goal was the complete mapping and understanding of all the genes of human beings.” - Human Genome Project.
Understanding human genome implies discovering and characterizing all the functional elements encoded within. Even though less than two percent of the mammalian genome corresponds to protein coding regions, it is estimated that about 5% of it is under purifying selection. This indicates that a majority of the regions under selection are non-coding regulatory elements or cis-regulatory elements. Genome wide association studies (GWAS) have identified mutations in non-coding regulatory regions as the most common features associated with human disease. Since many of these mutations affect elements that control the expression of key determinants of cell fate and phenotype, understanding how these mutations affect the networks that control gene expression opens the way for the development of new classes of drugs.
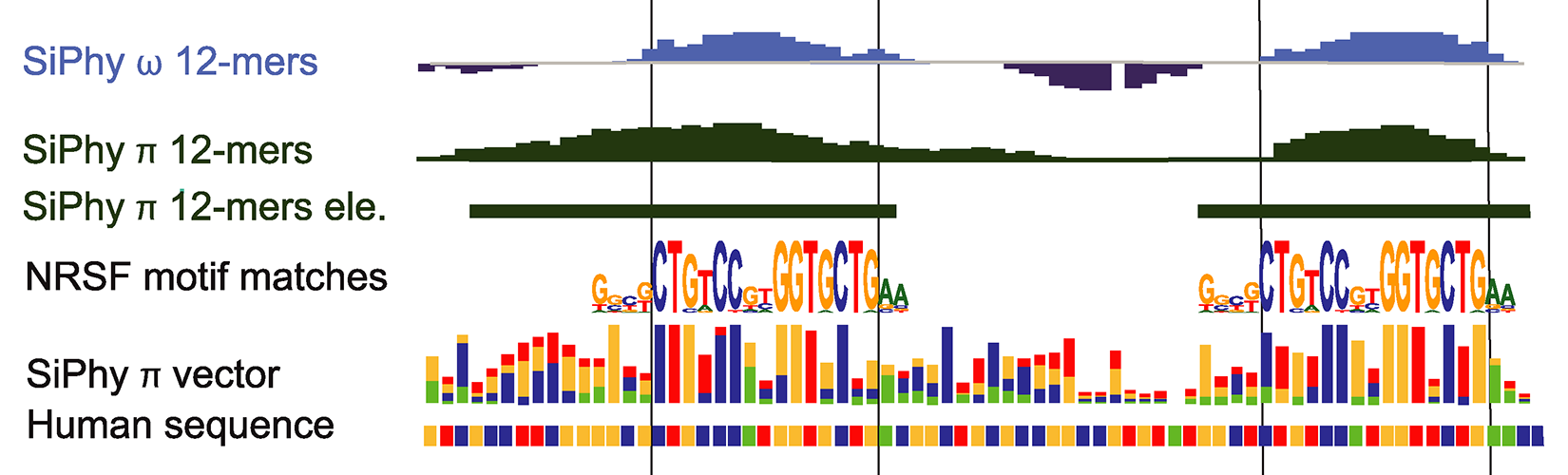
Most well-characterized enhancers are deeply conserved. In contrast, genome-wide comparative studies of steady-state systems showed that only a small fraction of active enhancers are conserved. To better understand conservation of enhancer activity, we recently used a comparative genomics approach that integrates temporal expression and epigenetic profiles in an innate immune system. We found that gene expression programs diverge among mildly induced genes, while being highly conserved for strongly induced genes. The fraction of conserved enhancers varies greatly across gene expression programs, with induced genes and early-response genes, in particular, being regulated by a higher fraction of conserved enhancers. Clustering of conserved accessible DNA sequences within enhancers identified shared sequence motifs (including motifs for known factors), as well as many with unknown function. We further show that the number of instances of these motifs is a strong predictor of the responsiveness of a gene to pathogen detection.