KRAS Inhibition
Characterize response and resistance to KRAS inhibition using in vivo RNAi and genome editing
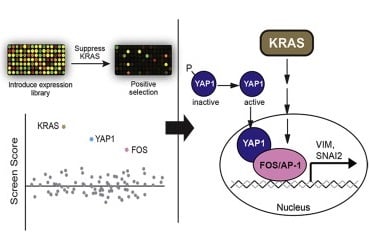
KRAS is mutated in >30% of lung cancer. While most studies in the literature use inducible Kras cDNA, conditional shRNA system allows knockdown of endogenous levels of Kras in vivo, offering a more physiologic model of Kras inhibition. We will model the response and resistance to Kras inhibition in mouse models.
Our initial observations showed that Kras-driven tumors relapse and become Kras-independent after long-term Kras inhibition (Shao, Xue, et al, 2014). We will explore the molecular basis of resistance to Kras inhibition. To completely delete the oncogenic KRAS gene and compare the phenotype with RNAi-mediated knockdown, we plan to use CRISPR to generate floxed KRAS conditional knockout alleles in a panel of cancer cell lines representing a variety of cancer types. These small RNA-based experiments will generate in vivo and in vitro platforms to uncover important KRAS biology. Read more