Development of chemical probes to study protein citrullination
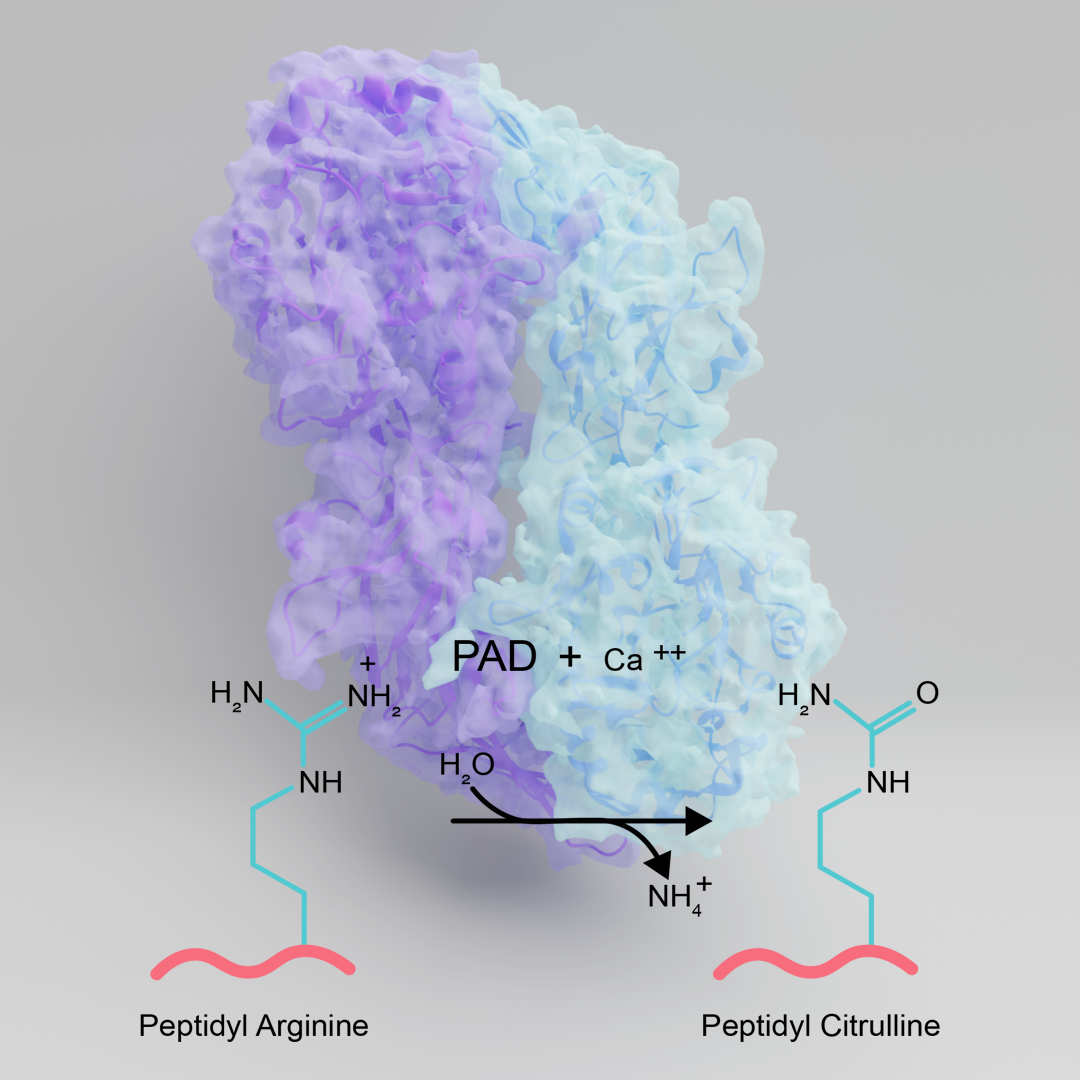
Protein citrullination is a post-translational modification that is catalyzed by the Protein Arginine Deiminases or PADs. This modification involves hydrolyzing the guanidinium group to form citrulline, a non-genetically encoded amino acid. Protein citrullination helps control numerous cell signaling pathways including the epigenetic regulation of gene expression, pluripotency, thrombotic events, and skin differentiation. In addition to these normal physiological functions, aberrantly increased protein citrullination is a hallmark of numerous inflammatory diseases including Rheumatoid Arthritis, Lupus, ALS, and cancer. To aid our understanding of the physiological roles of protein citrullination and develop potential therapeutics, our lab has been focused on developing chemical probes to study protein citrullination. In fact, we are world leaders in this area as we developed the first potent cell-active inhibitors of the PADs. Moreover, our group was the first to show that PAD inhibitors could work in vivo to ameliorate disease incidence and severity in multiple models of Rheumatoid Arthritis, Lupus, and sepsis. Current efforts are focused on developing isozyme selective PAD inhibitors (there are five isozymes in mammals) and to develop proteomic methodologies to unveil the full scope of proteins that are citrullinated by these enzymes. We are also performing cell-based studies to understand the impact of citrullination on muscle cell differentiation and determining how the enzymes are regulated in cells. One new area that we are focusing on is to determine structures of the PADs bound to known interacting proteins.
