Substrate Envelope
How is a viral protease able to recognize and cleave diverse substrates with high specificity? How can the protease mutate to avoid inhibition but still process these diverse substrates?
HIV-1 Protease: Molecular Basis for Resistance and Strategies to Avoid Resistance
Our research to answer to these questions for HIV-1 protease led to major breakthroughs in elucidating molecular mechanisms underlying selection of mutations in drug resistance:
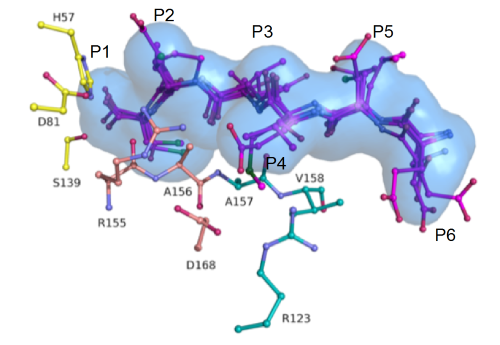
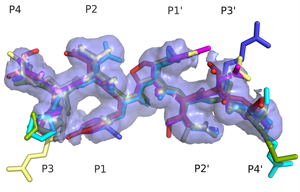 Superimposed HIV protease substrate complexes reveal the signature for substrate recognition: the substrate envelope. |
- HIV-1 protease recognizes its diverse substrate sequences through a conserved asymmetric shape that the substrates adopt. We defined this shape, or consensus volume, as the "substrate envelope".
- Active site resistance mutations within HIV-1 protease occur where the inhibitors protrude beyond the consensus substrate envelope and contact the protease. We discovered those positions are optimal for drug resistance to occur, as they are more important for inhibitor binding than for substrate binding.
- Protease inhibitors that fit within the substrate envelope are less susceptible to drug resistance – we discovered extremely potent inhibitors (single digit pM) that fit within the substrate envelope and validate our theory.
- We discovered the impact of mutations on inhibitor binding propagates throughout the active site through a highly cooperative network of interactions. Even the impact of distal mutations is propagated through this network (of hydrophobic sliding and intra-molecular interactions) to pivotal residues at the active site through alterations in the dynamic ensemble of the enzyme, thereby contributing to drug resistance.
- Resistance is further accentuated through co-evolution of the cleavage sites that mutate to conserve the substrate envelope shape.
In many HIV-1 protease variants, multiple site mutations co-evolve to both decrease the affinity of a particular inhibitor and increase the viability and fitness of the enzyme. We hypothesize that the impact of particular mutations on conferring drug resistance is not simply additive, but that these mutations have a complex interdependent effect leading to viable variants that are highly resistant to existing drugs.
All of our discoveries translate to elucidating the underlying molecular mechanisms of other quickly evolving diseases as do our strategies for avoiding drug resistance.
HCV NS3/4A Protease: Validation of Resistance Mechanisms and Design Strategies
We discovered that the "rules" that govern drug resistance in HIV-1 protease –both in terms of the substrate envelope and the dynamic network models– also apply to the viral protease of Hepatitis C (HCV), and explain genotypic differences. In this effort we have also become the leader of determining and elucidating the structures of inhibitor complexes with HCV NS3/4A protease, having determined more than 50 crystal structures of substrate and inhibitor complexes. We have synthesized more than 45 existing and novel inhibitors and performed more than 50 extensive molecular dynamics simulations. We collaborate extensively with both academic and industrial investigators. Currently we are focused on developing potent inhibitors to globally prevalent genotypes of HCV that not well inhibited by current drugs, in an attempt to avoid resistance among these patient populations.
Relevant Publications
Elucidating the Substrate Envelope of Enterovirus 68-3C Protease: Structural Basis of Specificity and Potential Resistance. Viruses. Azzolino VN, Shaqra AM, Ali A, Kurt Yilmaz N, Schiffer CA. . 2024 Sep 5;16(9):1419. doi: 10.3390/v16091419. PMID: 39339895; PMCID: PMC11437433.
Molecular Basis for Differential Patterns of Drug Resistance in Influenza N1 and N2 Neuraminidase.
Prachanronarong KL, Özen A, Thayer KM, Yilmaz LS, Zeldovich KB, Bolon DN, Kowalik TF, Jensen JD, Finberg RW, Wang JP, Kurt-Yilmaz N, Schiffer CA. J Chem Theory Comput. 2016 Dec 13;12(12):6098-6108. Epub 2016 Nov 17.
Drug resistance against HCV NS3/4A inhibitors is defined by the balance of substrate recognition versus inhibitor binding. Romano KP, Ali A, Royer WE, Schiffer CA.
Proc Natl Acad Sci U S A. 2010 Dec 7;107(49):20986-91. doi: 10.1073/pnas.1006370107. Epub 2010 Nov 17.