How do RNA-binding proteins control germline development and embryogenesis?
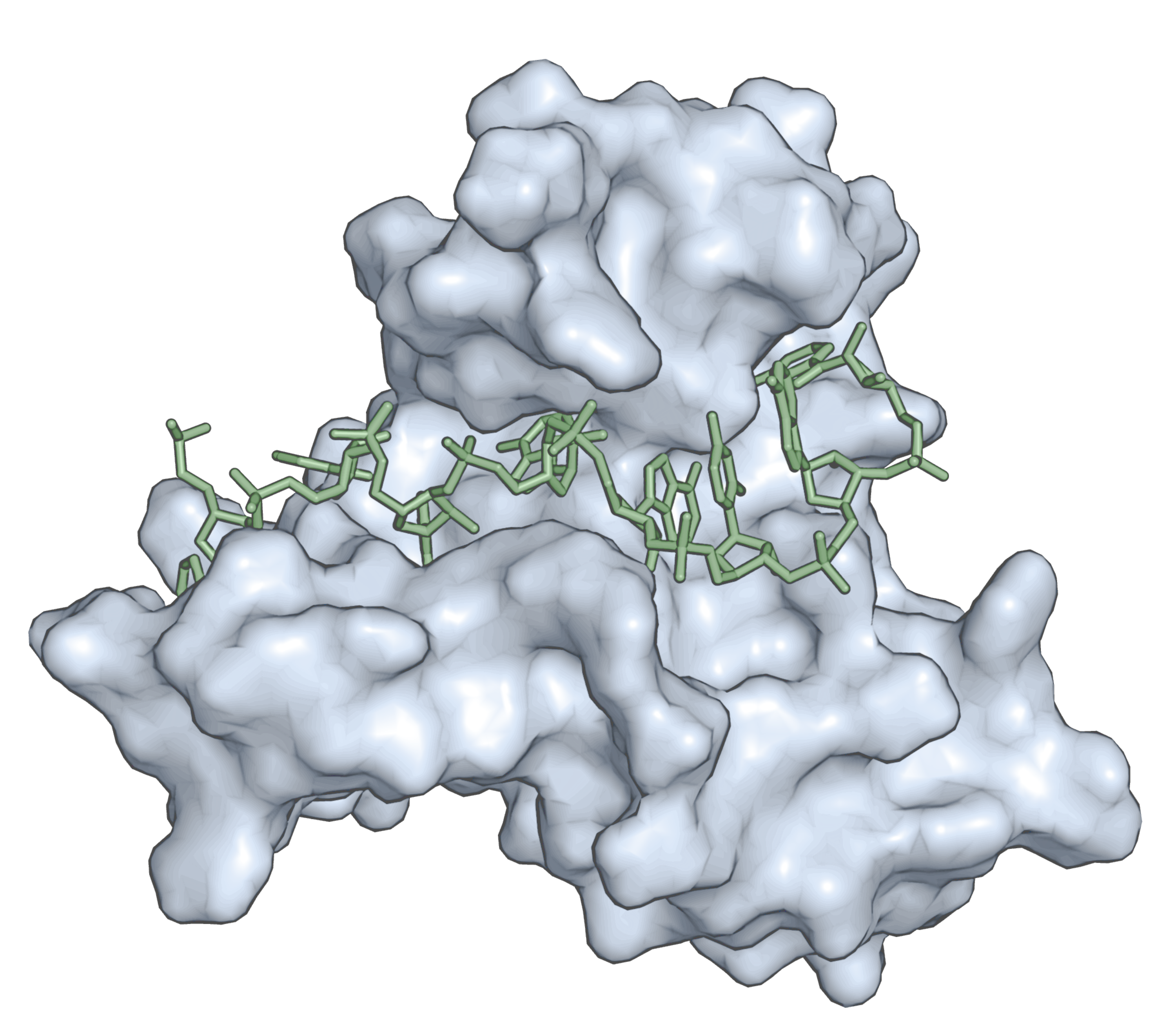 RNA-binding proteins (RBPs) govern each the major transitions in the germline and early embryo. These RBPs come from several different families that are highly conserved in mammals. In many cases, it remains unclear how the RNA binding properties contribute to gene regulation. For example, how do affinity, specificity, stability, cooperativity, and folding transitions contribute to phenotype? We are in a unique position to address these questions directly. We have expressed and purified many C. elegans RBPs over the course of the past ten years and have begun to dissect their biochemical properties using quantitative in vitro tools. Thanks to advances in genome editing, we now have the ability to introduce targeted mutations into the endogenous locus in C. elegans to determine phenotype.
RNA-binding proteins (RBPs) govern each the major transitions in the germline and early embryo. These RBPs come from several different families that are highly conserved in mammals. In many cases, it remains unclear how the RNA binding properties contribute to gene regulation. For example, how do affinity, specificity, stability, cooperativity, and folding transitions contribute to phenotype? We are in a unique position to address these questions directly. We have expressed and purified many C. elegans RBPs over the course of the past ten years and have begun to dissect their biochemical properties using quantitative in vitro tools. Thanks to advances in genome editing, we now have the ability to introduce targeted mutations into the endogenous locus in C. elegans to determine phenotype.
We use quantitative in vitro fluorescence-based electrophoretic mobility shift (F-EMSA) and polarization (FP) assays to monitor the kinetics and thermodynamics of RNA binding activity. In collaboration with Francesca Massi, we use NMR to evaluate the structure and dymanics of critical protein-RNA interactions. As such, we have a developing understanding of how structure relates to the biochemical properties of each protein. By contrast, we have limited understanding of how the structure and biochemical characteristics contribute to mutant phenotypes. Guided by our biochemical and structural insights, we are making precise mutations in endogenous RBPs and investigate the outcome in terms of phenotype, pattern of expression, and changes in downstream gene expression.
Recent Publications:
Tavella, D., Ertekin, A, Schaal, H, Ryder, S.P., and Massi, F, (2020) A disorder-to-order transition mediates RNA-binding of the Caenorhabditis elegans protein MEX-5. Biophys J. 118, 2001-2014.
Han, B., Antkowiak, K.R., Fan, X., Rutigliano, M., Ryder S.P., and Griffin, E.E., (2018) Polo-like kinase couples cytoplasmic protein gradients in the elegans zygote. Curr. Biol., 28(1):60-69.e8
Tamburino, A., Kaymak, E., Shrestha, S., Ryder, S.P., Walhout, A.J.M., (2017) PRIMA: a gene-centered, RNA-to-protein method for mapping RNA-protein interactions. Translation e1295130. doi: 10.1080/21690731.2017.1295130

