 |
|
| Thoru Pederson, PhD, and Hanhui Ma, PhD |
A new “app” for finding and mapping chromosomal loci using multicolored versions of CRISPR/Cas9, one of the hottest tools in biomedical research today, has been developed by scientists at UMass Medical School. This labeling system could be a key to understanding the spatial and temporal regulation of gene expression by allowing researchers to measure the precise linear distance between two known points on different chromosomes or two locations on the same chromosome in live human cells.
Detail of the findings were published in Proceedings of the National Academy of Sciences (PNAS) and first presented at the American Society for Cell Biology–International Federation for Cell Biology annual meeting in December.
The nucleus of every cell in our bodies (with the exception of gametes and red blood cells) must pack into it 23 pairs of chromosome—tight bundles of extremely long strands of DNA wound around protein knobs. For a gene to be transcribed and expressed, it must be accessible on the chromosome. Scientists have long suspected that the position of a chromosome within the nucleus affects gene accessibility and plays a critical role in everything from embryonic development to cancer.
Knowing the location and the intra-nuclear conformation of chromosomes is critical to understanding how genes actually work because the human cell nucleus is a very crowded place, according to study authors Thoru Pederson, PhD, the Vitold Arnett professor of cell biology and professor of biochemistry & molecular pharmacology, and research specialist Hanhui Ma, PhD.
 |
|
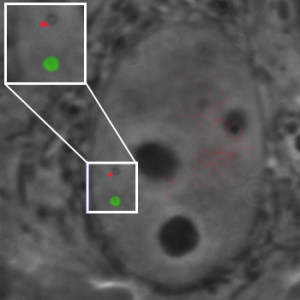
| Dual-color CRISPR labeling of two DNA sequences on chromosome 9 in a live human cell. |
By deploying pairs of fluorescent tags from their three-color system, Dr. Pederson and colleagues showed that it’s possible to plot where a chromosome is inside the cell nucleus and where it is in relation to other chromosomes. Their CRISPR app can also measure the distance between two points on the same chromosome, giving a read-out of chromosome compaction, which is a key factor in gene expression.
Precisely locating chromosomes in the nucleus of living cells has been a holy grail in cell biology since 1968, when Joseph Gall and Mary Lou Pardue first demonstrated the detection of specific loci. That discovery helped open the era of genetic testing, but the early techniques required fixed (i.e. dead) cells. In the intervening years, researchers have adapted new methods to probe live cells including transcription activator-like effectors (TALEs), which Ma and Pederson had recently introduced for lighting up genetic loci in living cells. But they subsequently came to believe that the rapidly emerging CRISPR system promised a more accurate map of a living nucleus and would be easier for scientists to employ.
Using their multicolored system, they were able to determine common locations for several chromosomes. Among their findings was that gene-rich chromosome 19 tends to be located in the middle of the nucleus, whereas gene-poor chromosome 18 is at the periphery. Also staying close to the center of the nucleus were chromosome 17 and five of the so-called acrocentric chromosomes, which have their distinctive centromeres close to the ends of one arm. One of the five acrocentrics is chromosome 21; the presence of an extra copy of chromosome 21 is called trisomy 21, and is the diagnostic marker for Down syndrome. Meanwhile, chromosomes 3 and 7 were at or close to the nuclear periphery.
The researchers were also able to distinguish the different locations for each diploid copy of the genes involved in organizing the nucleolus. The researchers say that they have plans to further tweak their two-color technique to study translocations—the abnormal switching of chromosomal segments—in human tumor cells.
Ultimately, the GPS app will provides scientists a new toolkit for “studying the 4D nucleome and the regulation of eukaryotic gene expression across a broad landscape of cell types and stages of development, differentiation and human disease,” the authors state in their publication.
The published study was funded in part by the U.S. National Science Foundation.