How mammalian SWI/SNF enzymes promote cell differentiation
Our research interests are focused on the biological functions of the mammalian SWI/SNF (mSWI/SNF) ATP-dependent chromatin remodeling enzymes. This enzyme family uses ATP hydrolysis to break histone-DNA contacts and alter nucleosome structure and/or positioning. Our work provided the first evidence that mammalian SWI/SNF enzymes remodel chromatin at endogenous gene loci, and we established that the mSWI/SNF enzymes are essential for many tissue-differentiation events. Our major efforts have centered on how SWI/SNF enzymes cooperate with tissue-specific transcription factors and histone modifying enzymes to promote tissue-specific gene expression, with an emphasis on myogenesis and adipogenesis. In particular, we have focused on connections between chromatin remodeling enzymes, arginine methyltransferases (PRMTs) and arginine deiminases (PADs) that are known chromatin modifying enzymes. Additional work has explored changes in higher-order gene organization that result from differentiation and signal transduction mechanisms that modify the activities of mSWI/SNF chromatin remodeling enzymes via post-translational modifications of the individual enzyme subunits.
Relevant Publications
Olea-Flores, M, T Sharma, O Verdejo-Torres, I DiBartolomeo, PR Thompson, T Padilla-Benavides, AN Imbalzano. 2024. Muscle-specific pyruvate kinase isoforms, Pkm1 and Pkm2, regulate mammalian SWI/SNF proteins and histone 3 phosphorylation during myoblast differentiation. FASEB J. 38:e23702. DOI:10.1096/fj.202400784R [PMCID: in process]
Sharma, T, DCL Robinson, H Witwicka, FJ Dilworth and AN Imbalzano. 2021. The Bromodomains of the mammalian SWI/SNF (mSWI/SNF) ATPases Brahma (BRM) and Brahma Related Gene 1 (BRG1) promote chromatin interaction and are critical for skeletal muscle differentiation. Nucleic Acids Res. 49:8060-8077. DOI:10.1093/nar/gkab617 [PMCID: PMC8373147]
Padilla-Benavides, T, DT Haokip, Y Yoon, P Reyes-Gutierrez, JA Rivera-Pérez and AN Imbalzano. 2020. CK2-dependent phosphorylation of the Brg1 chromatin remodeling enzyme occurs during mitosis. Int. J. Mol. Sci. 21:923. DOI:10.3390/ijms21030923 [PMCID: PMC7036769 ]
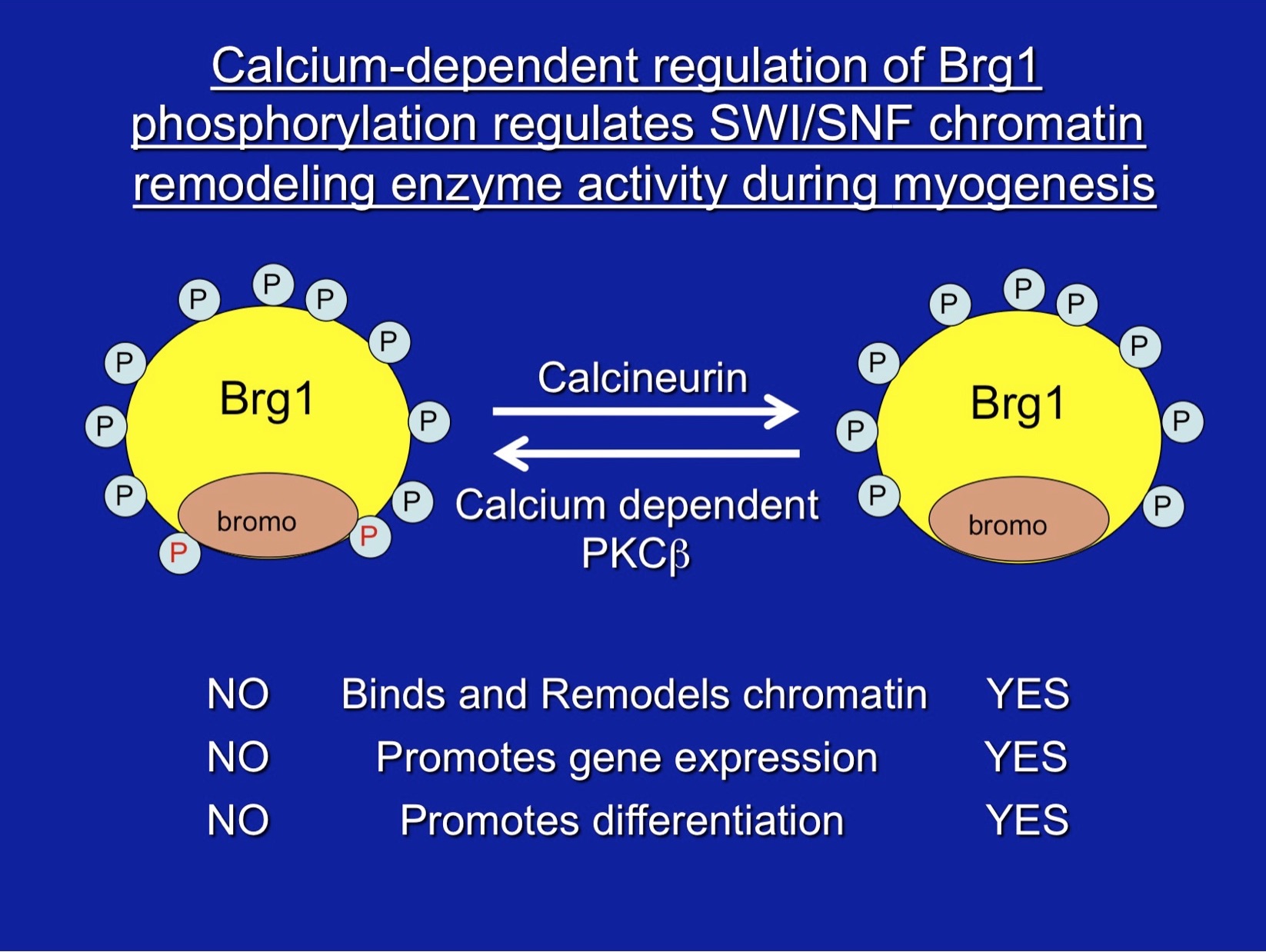
Padilla-Benavides, T, P Reyes-Gutierrez and AN Imbalzano. 2020. Regulation of mammalian SWI/SNF family of chromatin remodeling enzymes by phosphorylation during myogenesis. Biology 9:E152. DOI: 10.3390/biology9070152 [PMCID: PMC7407365] (Review)
Witwicka, H, J Nogami, SA Syed, K Maehara, T Padilla-Benavides, Y Ohkawa and AN Imbalzano. 2019. Calcineurin broadly regulates the initiation of skeletal muscle-specific gene expression by binding target promoters and facilitating the interaction of the SWI/SNF chromatin remodeling enzyme. Mol. Cell. Biol. 39: e00063-19. DOI: 10.1128/MCB.00063-19 [PMCID: PMC6751634]
Padilla-Benavides, T, BT Nasipak, AL Paskavitz, DT Haokip, JM Schnabl, JA Nickerson and AN Imbalzano. 2017. Casein kinase 2–mediated phosphorylation of Brahma-related gene 1 controls myoblast proliferation and contributes to SWI/SNF complex composition. J. Biol. Chem. 292:18592-18607. DOI: 10.1074/jbc.M117.799676 [PMCID: PMC5682968].
Harada, A, Y Ohkawa, and AN Imbalzano. 2017. Temporal regulation of chromatin during myoblast differentiation. Semin. Cell Dev. Biol. 72:77-86. DOI: 10.1016/j.semcdb.2017.10.022 [PMCID: PMC5723239]. (Review).
Barutcu, AR, BR Lajoie, AJ Fritz, RP McCord, JA Nickerson, AJ van Wijnen, JB Lian, JL Stein, J Dekker, GS Stein, AN Imbalzano. 2016. SMARCA4 regulates gene expression and higher-order chromatin structure in proliferating mammary epithelial cells. Genome Res. 26:1188-1201. DOI: 10.1101/gr.201624.115 [PMCID: PMC5052043]
LeBlanc, SE, Q Wu, P Lamba, S Sif, and AN Imbalzano. 2016. Promoter-enhancer looping at the PPARγ2 locus during adipogenic differentiation requires the Prmt5 methyltransferase. Nucleic Acids Res. 44:5133-5147. DOI: 10.1093/nar/gkw129 [PMCID: PMC4914087]
Nasipak, BT, T Padilla-Benavides, KM Green, JD Leszyk, W Mao, S Konda, S Sif, SA Shaffer, Y Ohkawa, and AN Imbalzano. 2015. Opposing calcium dependent signaling pathways control skeletal muscle differentiation by regulating a chromatin remodeling enzyme. Nature Commun. 6:7441 | DOI: 10.1038/ncomms8441 [PMCID: PMC4530624]
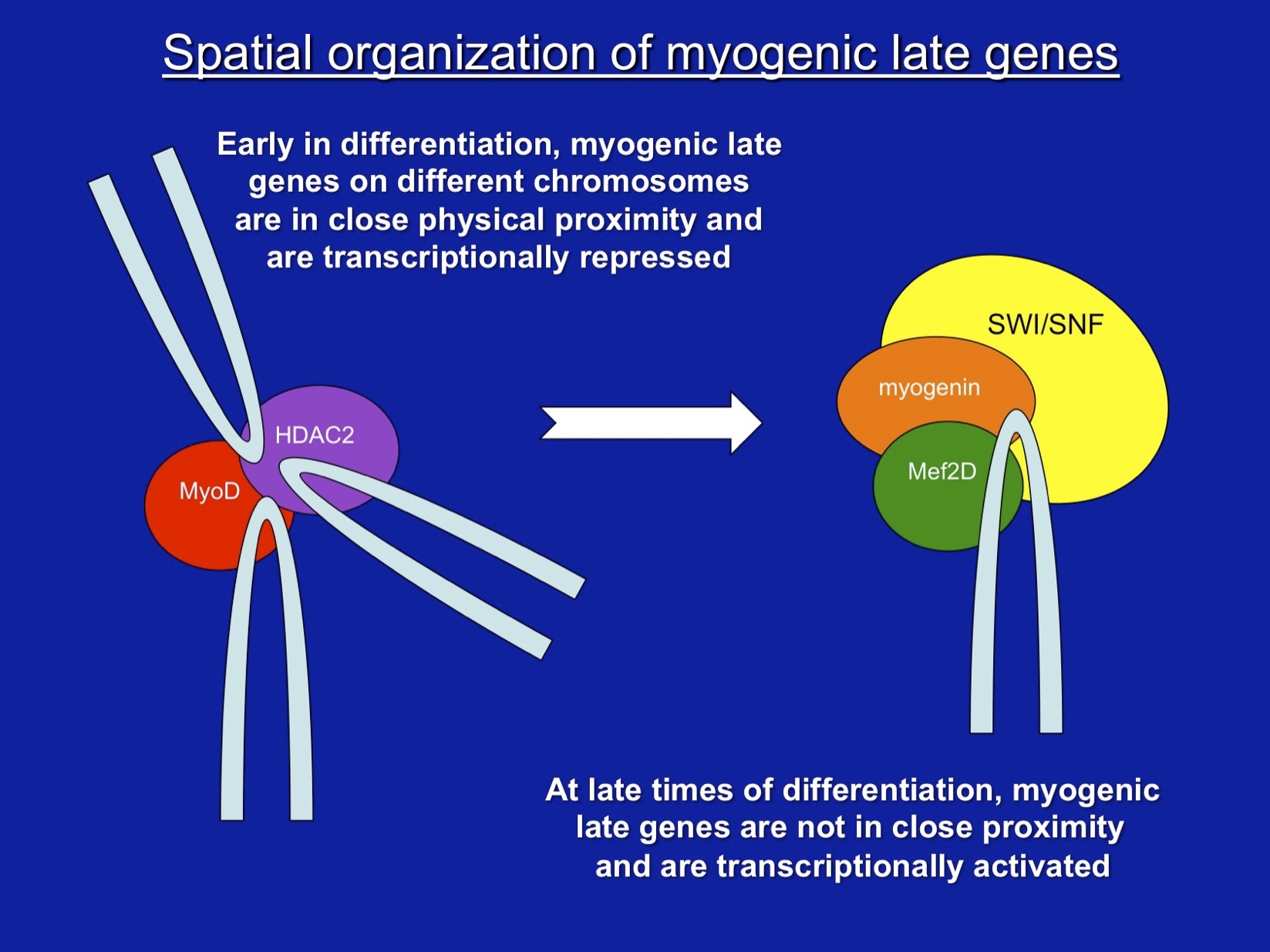
Harada, A, C Mallappa, S Okada, JT Butler, SP Baker, JB Lawrence, Y Ohkawa, and AN Imbalzano. 2015. Spatial re-organization of myogenic regulatory sequences temporally controls gene expression. Nucleic Acids Res. 43:2008-2021. DOI: 10.1093/nar/gkv046 [PMCID: PMC4344497]
LeBlanc, SE, Q Wu, AR Barutcu, H Xiao, Y Ohkawa, and AN Imbalzano. 2014. The PPARg locus makes long-range chromatin interactions with selected tissue-specific gene loci during adipocyte differentiation in a protein kinase A dependent manner. PLoS One 9(1): e86140. [PMCID: PMC3896465]
