Primer on Dr. Green's research
Michael Green, MD, PhD, stood as a towering figure in molecular biology and cancer research. His groundbreaking discoveries in gene regulation established an unparalleled standard for innovation, and his research illuminated the mechanics of gene transcription and paved the way for better therapeutics for cancer and rare diseases. His unexpected death in February 2023 left a gaping hole in the collective heart of the UMass Chan community.
This quote by John Quincy Adams aptly captures Dr. Green’s impact on us all: “If your actions inspire others to dream more, learn more, do more, and become more, you are a leader.”
Read along for a primer on his most influential research.
Mid-1980s: In vitro transcription
 While a postdoctoral fellow in Thomas Maniatis’ lab at Harvard, Dr. Green’s career got off to an illustrious start. At the time, scientists wishing to explore the structure and function of RNA needed to purify viral RNAs from living cells—a laborious and complex procedure. But there was a paradigm shift when the Maniatis lab developed a way to make RNA in a test tube from a DNA template. This pivotal methodology is at the root of countless other discoveries. “It’s the same technology that underlies the COVID vaccine and all mRNA therapeutics,” says Dr. Green’s former graduate student, Phillip Zamore, PhD, director of the RNA Therapeutics Institute at UMass Chan. With the simplification of RNA synthesis, scientists could readily study specific RNA molecules or genes of interest, shed light on genetic diseases, and streamline the study of gene transcription and translation.
While a postdoctoral fellow in Thomas Maniatis’ lab at Harvard, Dr. Green’s career got off to an illustrious start. At the time, scientists wishing to explore the structure and function of RNA needed to purify viral RNAs from living cells—a laborious and complex procedure. But there was a paradigm shift when the Maniatis lab developed a way to make RNA in a test tube from a DNA template. This pivotal methodology is at the root of countless other discoveries. “It’s the same technology that underlies the COVID vaccine and all mRNA therapeutics,” says Dr. Green’s former graduate student, Phillip Zamore, PhD, director of the RNA Therapeutics Institute at UMass Chan. With the simplification of RNA synthesis, scientists could readily study specific RNA molecules or genes of interest, shed light on genetic diseases, and streamline the study of gene transcription and translation.
Late 1980s-early 1990s: Splicing
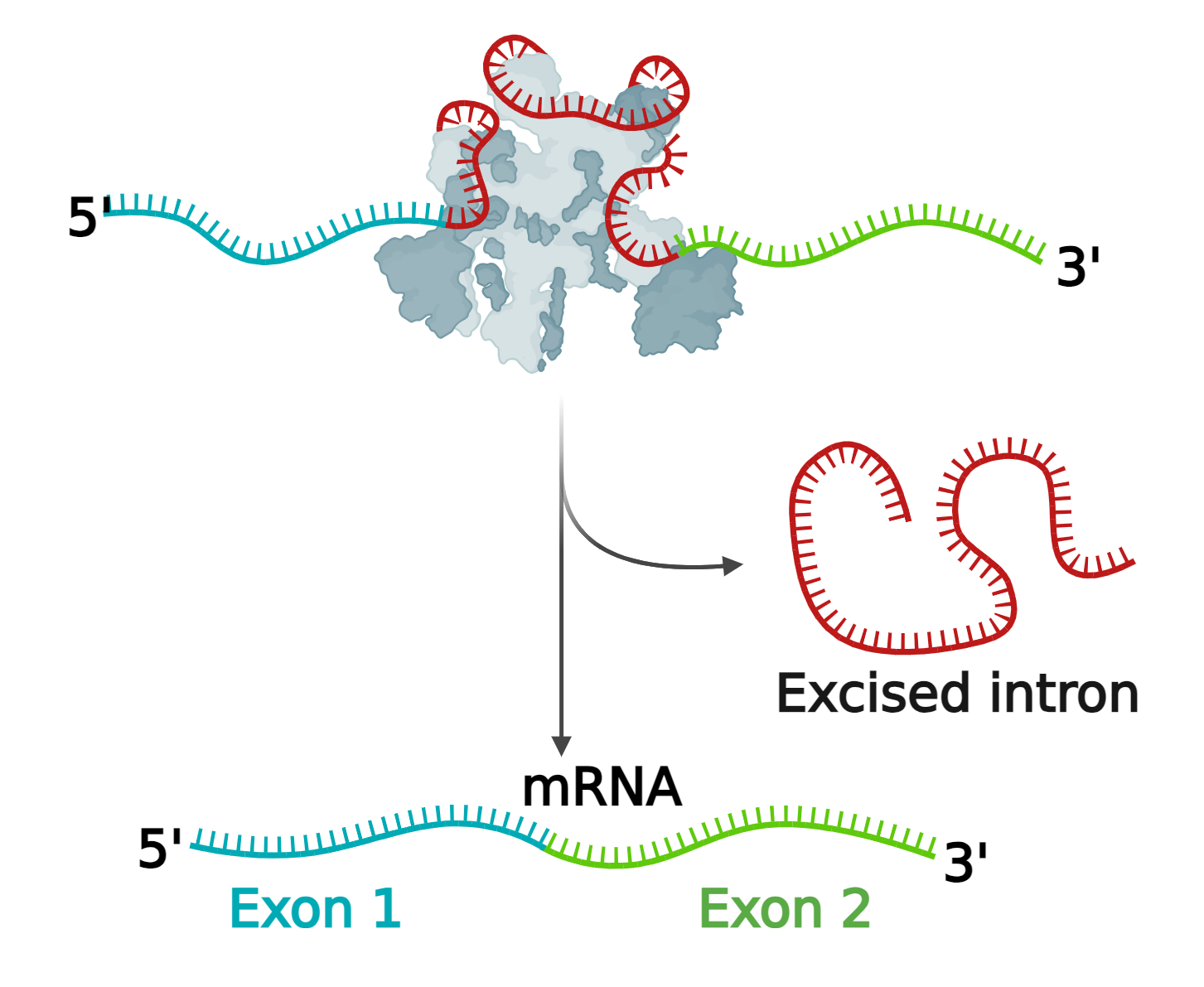 When Dr. Green was a newly-minted independent researcher, he set out to understand splicing–a crucial step in transcription in which only the sections of the RNA molecule that code for proteins are retained.
When Dr. Green was a newly-minted independent researcher, he set out to understand splicing–a crucial step in transcription in which only the sections of the RNA molecule that code for proteins are retained.
How does the cell “know” which RNA segments code for proteins, which do not, and what biochemical cues signal the boundaries between the two?
According to Dr. Zamore, Dr. Green accomplished this task by dividing his lab into two teams. One group identified the proteins that recognize coding sections of RNA (exons), non-coding sections (introns) and the borders between the two. The second group determined the sequences that guide the correct and efficient removal of introns and reconnect remaining exons. This work culminated in a major discovery describing crucial components of the biochemical machinery that came to be called the “spliceosome”. One of these proteins, U2AF, recognizes an intron-exon boundary and, when mutated, contributes to the development of myelodysplastic syndromes and acute myeloid leukemia.
Mechanisms of transcription activation
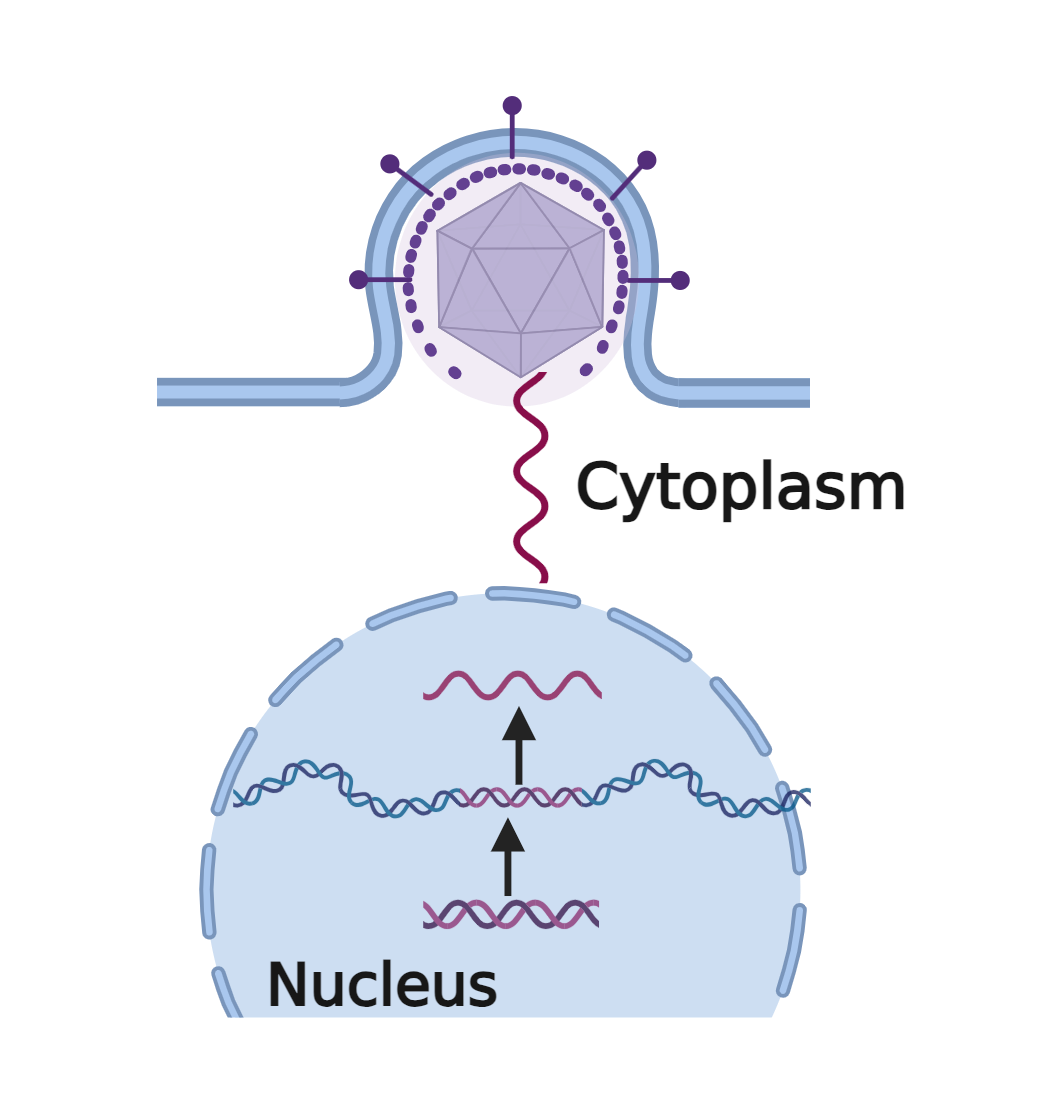 Dr. Green was successful in helping us understand how the RNA molecule is processed for the subsequent translation step, but he also wanted to unpack the molecular events needed to initiate the transcription process. He and former graduate student James Lillie, PhD, enlisted viruses to study the problem.
Dr. Green was successful in helping us understand how the RNA molecule is processed for the subsequent translation step, but he also wanted to unpack the molecular events needed to initiate the transcription process. He and former graduate student James Lillie, PhD, enlisted viruses to study the problem.
“The adenovirus E1A protein was one of the early model systems for transcriptional activation in eukaryotes [organisms with a complex cell structure] because we knew it turned on transcription of different genes in the virus and the host cell,” says Dr. Lillie.
What was missing was the “how”.
To find out, the two scientists deleted, added, fused, and otherwise manipulated individual segments of E1A for clues on how transcription works. Their experiments revealed that E1A had a modular structure–with a section that helps it gravitate toward the DNA and another segment that interacts with other protein complexes to initiate transcription. In doing so, E1A hijacks the transcription process in the host cell, forcing it to make viral genes. Dr. Green’s additional studies demonstrated, for the first time, that proteins—co-activators—are recruited to help stabilize the complex on the DNA to make transcription more efficient and accurate.
Together with his students, Dr. Green demonstrated that transcription is a team sport and they delineated each player’s role in achieving the final goal.
Mid-1990s: Chromatin remodeling
 Dr. Green was a master at breaking down seemingly impossible problems into manageable and solvable puzzles. After filling in many of the gaps of how transcription is activated and the RNA spliced, it was time to turn his attention to another problem: What happens if a gene needs transcribing, but the DNA configuration precludes it?
Dr. Green was a master at breaking down seemingly impossible problems into manageable and solvable puzzles. After filling in many of the gaps of how transcription is activated and the RNA spliced, it was time to turn his attention to another problem: What happens if a gene needs transcribing, but the DNA configuration precludes it?
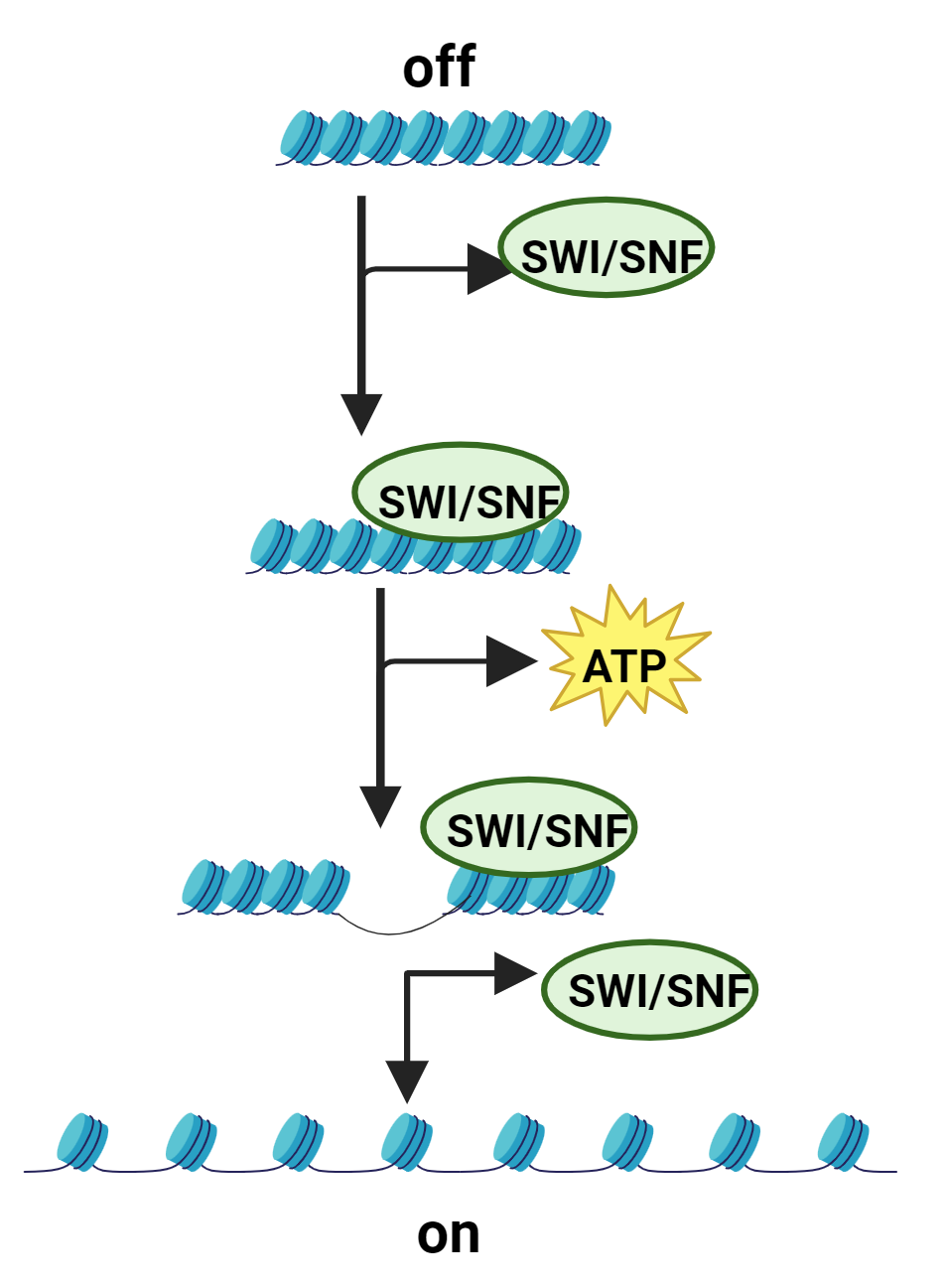
To fit in the nucleus, eukaryotic DNA is wrapped tightly around proteins called histones, forming nucleosomes. The nucleosomes are arranged like “beads on a string.” Collectively these components are referred to as chromatin. If the nucleosomes are too tightly wound (as in the figure to the left) the DNA may not be accessible to the RNA polymerase enzyme that needs to attach to it for transcription to occur.
How can this situation be rectified?
Scientists knew that a protein complex called SWItch/Sucrose Non-Fermentable (or SWI/SNF) activated transcription. What they didn’t know was how. Dr. Green and others jointly solved the mystery. They showed that SWI/SNF recruits adenosine triphosphate (ATP), using the energy to move the “beads on a string” and remodel the chromatin. These alterations allow the enzyme to bind to the DNA and transcription to occur.
“This work changed the way the field understood how the genome is organized and regulated,” says Thomas Fazzio, Professor of MCCB at UMass Chan.”
2000 and beyond: Epigenetic silencing and gene discovery
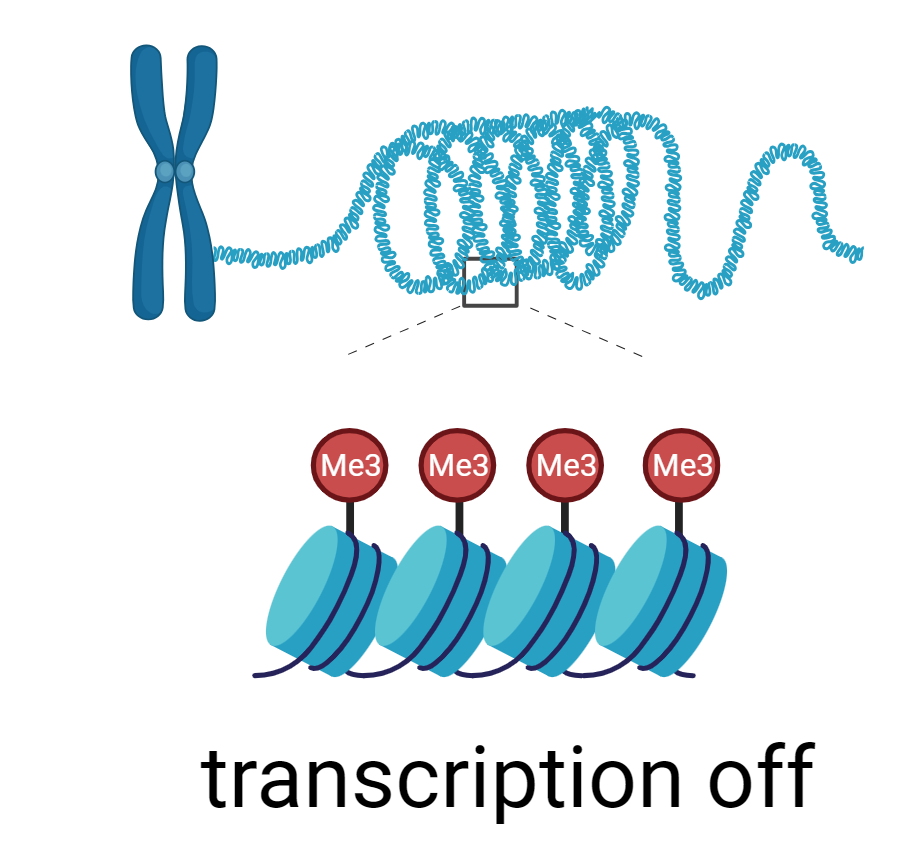 As translational research became the trend, Dr. Green found ways to contribute to the cause. He recognized that processes that lead to faulty transcription could trigger diseases. Gene silencing serves as a clear illustration. Cancer, for example, can arise when tumor suppressors are turned off. This inactivation can sometimes happen through a process known as 'epigenetic silencing,' which can occur when methyl groups are added to the DNA that wraps around the histones, or when acetyl groups are removed from the histones. Both modifications hide the gene from the gene transcription machinery.
As translational research became the trend, Dr. Green found ways to contribute to the cause. He recognized that processes that lead to faulty transcription could trigger diseases. Gene silencing serves as a clear illustration. Cancer, for example, can arise when tumor suppressors are turned off. This inactivation can sometimes happen through a process known as 'epigenetic silencing,' which can occur when methyl groups are added to the DNA that wraps around the histones, or when acetyl groups are removed from the histones. Both modifications hide the gene from the gene transcription machinery.
Scientists didn’t know whether epigenetic silencing occurred randomly or through an intricate pathway triggered by an oncogene.
To find out, Dr. Green designed RNA interference screens to systematically turn off a single gene at a time to identify which one(s) could reactivate the silenced tumor suppressor. In doing so, they discovered that oncoproteins can direct gene silencing through specific pathways. This finding ruled out the notion that epigenetic silencing was a random process. An added benefit—the genes found to restore expression in the tumor suppressor became potential targets for anticancer therapies.
Dr. Green’s research will undoubtedly have a profound and lasting impact on science and on all of us who knew and respected him. We miss him dearly, but we will honor his legacy by continuing to research and combat cancer.
All graphics created in Biorender.com
The Michael R. Green, MD, PhD, Award in Graduate Research
